
Saccharopolyspora flava
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia<
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
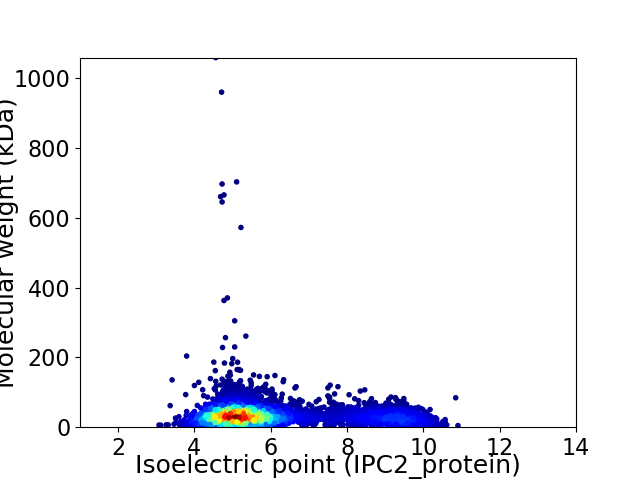
Virtual 2D-PAGE plot for 5595 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6TKP3|A0A1I6TKP3_9PSEU Sarcosine oxidase subunit delta OS=Saccharopolyspora flava OX=95161 GN=SAMN05660874_04016 PE=4 SV=1
MM1 pKa = 7.39LASGAQFGPIVIRR14 pKa = 11.84GNAPATGTSVTNTASVAGADD34 pKa = 3.63VDD36 pKa = 5.5SNTANNTASATTSLTAQAPALTVQKK61 pKa = 8.81TANPTSVTTAGAPIVYY77 pKa = 9.65SYY79 pKa = 10.92RR80 pKa = 11.84VSNIGNVPLTGISVAEE96 pKa = 4.04TAFTGTGPRR105 pKa = 11.84PSASCPATSLDD116 pKa = 3.67PGASMTCTANYY127 pKa = 7.57TATQADD133 pKa = 4.0VDD135 pKa = 4.11RR136 pKa = 11.84GSIVNTAVANGTSPAGVAVVSPPTTATVTAAASPSLTVAKK176 pKa = 9.03TASPSDD182 pKa = 3.32AASFTVGRR190 pKa = 11.84VITYY194 pKa = 9.1SYY196 pKa = 10.8RR197 pKa = 11.84ITNTGNVTLTNVRR210 pKa = 11.84PTDD213 pKa = 3.39TAFNGSGPRR222 pKa = 11.84PTPTCPSGAASLAPGASVTCTATYY246 pKa = 9.53TVTQADD252 pKa = 3.77VDD254 pKa = 4.24RR255 pKa = 11.84GSLTNTATATGTPPSGSSPPTSPPSTVTVPGDD287 pKa = 3.49SAPALTVRR295 pKa = 11.84KK296 pKa = 9.15SASPTTMNAAGNTITYY312 pKa = 9.97RR313 pKa = 11.84FLVTNTGNVTLNSVSIAEE331 pKa = 4.16PTFTGTGGAPAVTCPSGPLLPGAQTTCTATYY362 pKa = 10.67SVTQADD368 pKa = 4.27MDD370 pKa = 4.3RR371 pKa = 11.84GTIDD375 pKa = 3.2NTATASAVAPDD386 pKa = 3.62ATRR389 pKa = 11.84VTSAPSSARR398 pKa = 11.84VTADD402 pKa = 3.7RR403 pKa = 11.84APALAVVKK411 pKa = 10.33SATPTTVTAAGDD423 pKa = 3.49AVNYY427 pKa = 9.09SFRR430 pKa = 11.84VTNTGNVSLDD440 pKa = 3.82TITVADD446 pKa = 4.04TQFSGTGTPPAPSCPPGVLAPGASTTCTASYY477 pKa = 8.51TATQADD483 pKa = 3.73IDD485 pKa = 4.35AGSISNTATASGRR498 pKa = 11.84DD499 pKa = 3.67PTTGNGTTSPPSSATVIADD518 pKa = 3.67ATPGLSLVKK527 pKa = 9.56TADD530 pKa = 3.51PTRR533 pKa = 11.84VTAAGQTVTFGFLVTNTSAVTLNDD557 pKa = 3.58PQIAEE562 pKa = 4.1TDD564 pKa = 4.02FTGSGPAPAPTCPSGPLAPGAQVRR588 pKa = 11.84CTATYY593 pKa = 7.36TTTQADD599 pKa = 3.56VDD601 pKa = 3.92RR602 pKa = 11.84GSIVNIATASATPPSGSPVVSDD624 pKa = 4.01PSTVTVTAAATPGITVVKK642 pKa = 8.82TATPQSVSAAGQTVDD657 pKa = 4.25YY658 pKa = 10.45AFQVTNTGNVTMNSVGITDD677 pKa = 3.84TEE679 pKa = 4.4FTGTGPAPEE688 pKa = 4.4VSCPSSTLAPGISMTCTASYY708 pKa = 8.5TATQADD714 pKa = 3.71IDD716 pKa = 4.18AGGFDD721 pKa = 3.82NTATATGTPPGSTPQTSPPSSTTVTAEE748 pKa = 3.94RR749 pKa = 11.84AASLALVKK757 pKa = 10.7SADD760 pKa = 3.66PSVVNAAGQTVDD772 pKa = 3.5YY773 pKa = 10.71SFLVTNTGNATVNDD787 pKa = 4.19LAVTEE792 pKa = 4.47TDD794 pKa = 3.95FSGSGPAPEE803 pKa = 4.84VSCPTDD809 pKa = 3.4PLAPGASTTCTASYY823 pKa = 10.79SATQADD829 pKa = 3.87IDD831 pKa = 4.4AGSITNTATAGGTDD845 pKa = 3.64PAGDD849 pKa = 4.08PVTSPPSSATVVAAVEE865 pKa = 4.26GDD867 pKa = 2.91ISLFKK872 pKa = 10.59SANPEE877 pKa = 4.07TVSAAGDD884 pKa = 3.4QVTFEE889 pKa = 4.18FLVTNVEE896 pKa = 4.12NVTLNAIRR904 pKa = 11.84VDD906 pKa = 3.64EE907 pKa = 4.44VEE909 pKa = 4.18FTGTGTLSPITCPPGPLAPGEE930 pKa = 4.1QRR932 pKa = 11.84TCTATYY938 pKa = 10.42AATQADD944 pKa = 4.1INAGSVTNTATASGTEE960 pKa = 3.9PDD962 pKa = 3.85GTDD965 pKa = 3.27VTSEE969 pKa = 3.95PASVVVTAEE978 pKa = 3.95AAPAMTVVKK987 pKa = 10.09SAQPQTVTAAGQTIDD1002 pKa = 3.85YY1003 pKa = 9.67SYY1005 pKa = 11.5QVTNTGNVTVDD1016 pKa = 3.39GVSVTEE1022 pKa = 4.0TDD1024 pKa = 3.77FTGTGPAPTPSCPDD1038 pKa = 3.12GPLAPGEE1045 pKa = 4.48STTCTASYY1053 pKa = 8.51TATQADD1059 pKa = 3.85IDD1061 pKa = 4.12AGGITNTATTTGTPPGGPPVTSPPSTSTVTTDD1093 pKa = 3.52PSPALTVAKK1102 pKa = 9.21TGEE1105 pKa = 4.17PATVSAAGQTITYY1118 pKa = 9.33SYY1120 pKa = 10.63EE1121 pKa = 3.6VRR1123 pKa = 11.84NTGNQTVTDD1132 pKa = 3.93VAVDD1136 pKa = 3.36EE1137 pKa = 4.94SGFTGTGPAPAATCPTTTLAPGEE1160 pKa = 4.54STTCTATYY1168 pKa = 9.46TVTQGDD1174 pKa = 3.57IDD1176 pKa = 4.05AGGVTNTGVATAQPPGGAPPVSSPPSTVTTGVEE1209 pKa = 4.27PNPSLAVVKK1218 pKa = 10.44SATPTHH1224 pKa = 5.88VARR1227 pKa = 11.84AGDD1230 pKa = 3.68VITYY1234 pKa = 9.98HH1235 pKa = 6.84FEE1237 pKa = 3.78ATNTGNVTLTGVRR1250 pKa = 11.84LDD1252 pKa = 3.65EE1253 pKa = 4.45AAFTGTGDD1261 pKa = 4.79LGPATCTGGPTLAPGEE1277 pKa = 4.14KK1278 pKa = 9.32RR1279 pKa = 11.84ICDD1282 pKa = 3.48LEE1284 pKa = 4.29YY1285 pKa = 10.87AVTQADD1291 pKa = 3.33IDD1293 pKa = 3.98AGSISNGATASGQPPSGARR1312 pKa = 11.84VTSPPAFATVTSDD1325 pKa = 3.42PAPAIMVDD1333 pKa = 3.41KK1334 pKa = 10.13TADD1337 pKa = 3.48PTTLTGAGQTVTYY1350 pKa = 10.29SFLVTNSGNVTLHH1363 pKa = 6.67DD1364 pKa = 3.51VGIRR1368 pKa = 11.84EE1369 pKa = 4.37TGFSGSGQLPPANCPTTSLEE1389 pKa = 4.18PGQQTTCTTSYY1400 pKa = 11.07AAAQTDD1406 pKa = 3.45VDD1408 pKa = 3.74QGFIANSAVAVGTGPGGAEE1427 pKa = 3.69AMSEE1431 pKa = 3.82QSAARR1436 pKa = 11.84VEE1438 pKa = 4.67IPPQPALSLVKK1449 pKa = 10.18TSDD1452 pKa = 3.23PTTASAAGDD1461 pKa = 3.44QVTFEE1466 pKa = 4.3YY1467 pKa = 10.9AVTNTGNVTLNGVGVGEE1484 pKa = 4.54TEE1486 pKa = 4.39FSGTGQPPVPTCPPEE1501 pKa = 4.26AAVLAPGATVTCTASYY1517 pKa = 9.83TLTQADD1523 pKa = 3.55VDD1525 pKa = 4.18AGGTTNTAVAGATPPGGPPAVISPPSTSTVTVDD1558 pKa = 3.71PGPGLSVVKK1567 pKa = 10.22SANPSSVDD1575 pKa = 3.27SAGDD1579 pKa = 3.38VVTYY1583 pKa = 9.82EE1584 pKa = 4.03FVVTNTGNVTMEE1596 pKa = 4.31DD1597 pKa = 3.42VSVAEE1602 pKa = 4.24TDD1604 pKa = 3.85FTGTGPAPVATCPPGPLAPGAQTTCTAPYY1633 pKa = 9.43TVTQADD1639 pKa = 3.34VDD1641 pKa = 3.9AGSISNTATATGEE1654 pKa = 4.26PPGSDD1659 pKa = 3.1TPIPPSPPSSTTVTIDD1675 pKa = 3.39PAPGLSVVKK1684 pKa = 10.53SVDD1687 pKa = 3.26PAGPEE1692 pKa = 3.99SFAVGQEE1699 pKa = 3.75LTYY1702 pKa = 10.9TFVATNTGNVTLHH1715 pKa = 6.93DD1716 pKa = 3.92VAIDD1720 pKa = 3.99DD1721 pKa = 4.48EE1722 pKa = 4.88DD1723 pKa = 4.91FSGSGGLGEE1732 pKa = 4.41PTCTGDD1738 pKa = 3.66PASVAPGEE1746 pKa = 4.38QIVCTATYY1754 pKa = 10.8VVTQADD1760 pKa = 3.3VDD1762 pKa = 3.85AGGVRR1767 pKa = 11.84NSATATGIPPAPGSPPVTSPPSQVEE1792 pKa = 4.64VPQDD1796 pKa = 3.73PQPGLSLVKK1805 pKa = 9.76TADD1808 pKa = 3.77TEE1810 pKa = 4.34HH1811 pKa = 6.5LTGPGQVITYY1821 pKa = 10.16SFLITNTGNVTLDD1834 pKa = 3.09RR1835 pKa = 11.84VAVDD1839 pKa = 3.19EE1840 pKa = 5.09GEE1842 pKa = 4.39FTGSGPLSPVTCPDD1856 pKa = 3.18EE1857 pKa = 5.22AGSLSPGEE1865 pKa = 4.15QATCTATYY1873 pKa = 9.45TVTQSDD1879 pKa = 3.65VDD1881 pKa = 3.87RR1882 pKa = 11.84GEE1884 pKa = 4.27LTNTATAGGTPPGGGGTTSPPSSATITGDD1913 pKa = 3.19AKK1915 pKa = 10.74PVLEE1919 pKa = 4.25VLKK1922 pKa = 9.21TAHH1925 pKa = 6.47WVDD1928 pKa = 4.12DD1929 pKa = 4.15NGSGVVDD1936 pKa = 4.64LGDD1939 pKa = 4.45RR1940 pKa = 11.84IAWTITATNTGGATATDD1957 pKa = 4.02VVVSDD1962 pKa = 4.06PTAGAVTCPATTLAPGEE1979 pKa = 4.64SMDD1982 pKa = 4.24CTAPDD1987 pKa = 3.69HH1988 pKa = 6.74VVDD1991 pKa = 4.37EE1992 pKa = 4.98ADD1994 pKa = 3.36VAAGEE1999 pKa = 4.34VANTAIATGRR2009 pKa = 11.84GPGGDD2014 pKa = 3.73PVTSPPASSTVEE2026 pKa = 3.82VGRR2029 pKa = 11.84PGTPPGPQPEE2039 pKa = 4.45EE2040 pKa = 3.8PGGPGDD2046 pKa = 3.98LATTGRR2052 pKa = 11.84DD2053 pKa = 3.22VGLLLALAGALLAFGGLLFSGARR2076 pKa = 11.84RR2077 pKa = 11.84RR2078 pKa = 11.84RR2079 pKa = 11.84RR2080 pKa = 11.84SS2081 pKa = 3.02
MM1 pKa = 7.39LASGAQFGPIVIRR14 pKa = 11.84GNAPATGTSVTNTASVAGADD34 pKa = 3.63VDD36 pKa = 5.5SNTANNTASATTSLTAQAPALTVQKK61 pKa = 8.81TANPTSVTTAGAPIVYY77 pKa = 9.65SYY79 pKa = 10.92RR80 pKa = 11.84VSNIGNVPLTGISVAEE96 pKa = 4.04TAFTGTGPRR105 pKa = 11.84PSASCPATSLDD116 pKa = 3.67PGASMTCTANYY127 pKa = 7.57TATQADD133 pKa = 4.0VDD135 pKa = 4.11RR136 pKa = 11.84GSIVNTAVANGTSPAGVAVVSPPTTATVTAAASPSLTVAKK176 pKa = 9.03TASPSDD182 pKa = 3.32AASFTVGRR190 pKa = 11.84VITYY194 pKa = 9.1SYY196 pKa = 10.8RR197 pKa = 11.84ITNTGNVTLTNVRR210 pKa = 11.84PTDD213 pKa = 3.39TAFNGSGPRR222 pKa = 11.84PTPTCPSGAASLAPGASVTCTATYY246 pKa = 9.53TVTQADD252 pKa = 3.77VDD254 pKa = 4.24RR255 pKa = 11.84GSLTNTATATGTPPSGSSPPTSPPSTVTVPGDD287 pKa = 3.49SAPALTVRR295 pKa = 11.84KK296 pKa = 9.15SASPTTMNAAGNTITYY312 pKa = 9.97RR313 pKa = 11.84FLVTNTGNVTLNSVSIAEE331 pKa = 4.16PTFTGTGGAPAVTCPSGPLLPGAQTTCTATYY362 pKa = 10.67SVTQADD368 pKa = 4.27MDD370 pKa = 4.3RR371 pKa = 11.84GTIDD375 pKa = 3.2NTATASAVAPDD386 pKa = 3.62ATRR389 pKa = 11.84VTSAPSSARR398 pKa = 11.84VTADD402 pKa = 3.7RR403 pKa = 11.84APALAVVKK411 pKa = 10.33SATPTTVTAAGDD423 pKa = 3.49AVNYY427 pKa = 9.09SFRR430 pKa = 11.84VTNTGNVSLDD440 pKa = 3.82TITVADD446 pKa = 4.04TQFSGTGTPPAPSCPPGVLAPGASTTCTASYY477 pKa = 8.51TATQADD483 pKa = 3.73IDD485 pKa = 4.35AGSISNTATASGRR498 pKa = 11.84DD499 pKa = 3.67PTTGNGTTSPPSSATVIADD518 pKa = 3.67ATPGLSLVKK527 pKa = 9.56TADD530 pKa = 3.51PTRR533 pKa = 11.84VTAAGQTVTFGFLVTNTSAVTLNDD557 pKa = 3.58PQIAEE562 pKa = 4.1TDD564 pKa = 4.02FTGSGPAPAPTCPSGPLAPGAQVRR588 pKa = 11.84CTATYY593 pKa = 7.36TTTQADD599 pKa = 3.56VDD601 pKa = 3.92RR602 pKa = 11.84GSIVNIATASATPPSGSPVVSDD624 pKa = 4.01PSTVTVTAAATPGITVVKK642 pKa = 8.82TATPQSVSAAGQTVDD657 pKa = 4.25YY658 pKa = 10.45AFQVTNTGNVTMNSVGITDD677 pKa = 3.84TEE679 pKa = 4.4FTGTGPAPEE688 pKa = 4.4VSCPSSTLAPGISMTCTASYY708 pKa = 8.5TATQADD714 pKa = 3.71IDD716 pKa = 4.18AGGFDD721 pKa = 3.82NTATATGTPPGSTPQTSPPSSTTVTAEE748 pKa = 3.94RR749 pKa = 11.84AASLALVKK757 pKa = 10.7SADD760 pKa = 3.66PSVVNAAGQTVDD772 pKa = 3.5YY773 pKa = 10.71SFLVTNTGNATVNDD787 pKa = 4.19LAVTEE792 pKa = 4.47TDD794 pKa = 3.95FSGSGPAPEE803 pKa = 4.84VSCPTDD809 pKa = 3.4PLAPGASTTCTASYY823 pKa = 10.79SATQADD829 pKa = 3.87IDD831 pKa = 4.4AGSITNTATAGGTDD845 pKa = 3.64PAGDD849 pKa = 4.08PVTSPPSSATVVAAVEE865 pKa = 4.26GDD867 pKa = 2.91ISLFKK872 pKa = 10.59SANPEE877 pKa = 4.07TVSAAGDD884 pKa = 3.4QVTFEE889 pKa = 4.18FLVTNVEE896 pKa = 4.12NVTLNAIRR904 pKa = 11.84VDD906 pKa = 3.64EE907 pKa = 4.44VEE909 pKa = 4.18FTGTGTLSPITCPPGPLAPGEE930 pKa = 4.1QRR932 pKa = 11.84TCTATYY938 pKa = 10.42AATQADD944 pKa = 4.1INAGSVTNTATASGTEE960 pKa = 3.9PDD962 pKa = 3.85GTDD965 pKa = 3.27VTSEE969 pKa = 3.95PASVVVTAEE978 pKa = 3.95AAPAMTVVKK987 pKa = 10.09SAQPQTVTAAGQTIDD1002 pKa = 3.85YY1003 pKa = 9.67SYY1005 pKa = 11.5QVTNTGNVTVDD1016 pKa = 3.39GVSVTEE1022 pKa = 4.0TDD1024 pKa = 3.77FTGTGPAPTPSCPDD1038 pKa = 3.12GPLAPGEE1045 pKa = 4.48STTCTASYY1053 pKa = 8.51TATQADD1059 pKa = 3.85IDD1061 pKa = 4.12AGGITNTATTTGTPPGGPPVTSPPSTSTVTTDD1093 pKa = 3.52PSPALTVAKK1102 pKa = 9.21TGEE1105 pKa = 4.17PATVSAAGQTITYY1118 pKa = 9.33SYY1120 pKa = 10.63EE1121 pKa = 3.6VRR1123 pKa = 11.84NTGNQTVTDD1132 pKa = 3.93VAVDD1136 pKa = 3.36EE1137 pKa = 4.94SGFTGTGPAPAATCPTTTLAPGEE1160 pKa = 4.54STTCTATYY1168 pKa = 9.46TVTQGDD1174 pKa = 3.57IDD1176 pKa = 4.05AGGVTNTGVATAQPPGGAPPVSSPPSTVTTGVEE1209 pKa = 4.27PNPSLAVVKK1218 pKa = 10.44SATPTHH1224 pKa = 5.88VARR1227 pKa = 11.84AGDD1230 pKa = 3.68VITYY1234 pKa = 9.98HH1235 pKa = 6.84FEE1237 pKa = 3.78ATNTGNVTLTGVRR1250 pKa = 11.84LDD1252 pKa = 3.65EE1253 pKa = 4.45AAFTGTGDD1261 pKa = 4.79LGPATCTGGPTLAPGEE1277 pKa = 4.14KK1278 pKa = 9.32RR1279 pKa = 11.84ICDD1282 pKa = 3.48LEE1284 pKa = 4.29YY1285 pKa = 10.87AVTQADD1291 pKa = 3.33IDD1293 pKa = 3.98AGSISNGATASGQPPSGARR1312 pKa = 11.84VTSPPAFATVTSDD1325 pKa = 3.42PAPAIMVDD1333 pKa = 3.41KK1334 pKa = 10.13TADD1337 pKa = 3.48PTTLTGAGQTVTYY1350 pKa = 10.29SFLVTNSGNVTLHH1363 pKa = 6.67DD1364 pKa = 3.51VGIRR1368 pKa = 11.84EE1369 pKa = 4.37TGFSGSGQLPPANCPTTSLEE1389 pKa = 4.18PGQQTTCTTSYY1400 pKa = 11.07AAAQTDD1406 pKa = 3.45VDD1408 pKa = 3.74QGFIANSAVAVGTGPGGAEE1427 pKa = 3.69AMSEE1431 pKa = 3.82QSAARR1436 pKa = 11.84VEE1438 pKa = 4.67IPPQPALSLVKK1449 pKa = 10.18TSDD1452 pKa = 3.23PTTASAAGDD1461 pKa = 3.44QVTFEE1466 pKa = 4.3YY1467 pKa = 10.9AVTNTGNVTLNGVGVGEE1484 pKa = 4.54TEE1486 pKa = 4.39FSGTGQPPVPTCPPEE1501 pKa = 4.26AAVLAPGATVTCTASYY1517 pKa = 9.83TLTQADD1523 pKa = 3.55VDD1525 pKa = 4.18AGGTTNTAVAGATPPGGPPAVISPPSTSTVTVDD1558 pKa = 3.71PGPGLSVVKK1567 pKa = 10.22SANPSSVDD1575 pKa = 3.27SAGDD1579 pKa = 3.38VVTYY1583 pKa = 9.82EE1584 pKa = 4.03FVVTNTGNVTMEE1596 pKa = 4.31DD1597 pKa = 3.42VSVAEE1602 pKa = 4.24TDD1604 pKa = 3.85FTGTGPAPVATCPPGPLAPGAQTTCTAPYY1633 pKa = 9.43TVTQADD1639 pKa = 3.34VDD1641 pKa = 3.9AGSISNTATATGEE1654 pKa = 4.26PPGSDD1659 pKa = 3.1TPIPPSPPSSTTVTIDD1675 pKa = 3.39PAPGLSVVKK1684 pKa = 10.53SVDD1687 pKa = 3.26PAGPEE1692 pKa = 3.99SFAVGQEE1699 pKa = 3.75LTYY1702 pKa = 10.9TFVATNTGNVTLHH1715 pKa = 6.93DD1716 pKa = 3.92VAIDD1720 pKa = 3.99DD1721 pKa = 4.48EE1722 pKa = 4.88DD1723 pKa = 4.91FSGSGGLGEE1732 pKa = 4.41PTCTGDD1738 pKa = 3.66PASVAPGEE1746 pKa = 4.38QIVCTATYY1754 pKa = 10.8VVTQADD1760 pKa = 3.3VDD1762 pKa = 3.85AGGVRR1767 pKa = 11.84NSATATGIPPAPGSPPVTSPPSQVEE1792 pKa = 4.64VPQDD1796 pKa = 3.73PQPGLSLVKK1805 pKa = 9.76TADD1808 pKa = 3.77TEE1810 pKa = 4.34HH1811 pKa = 6.5LTGPGQVITYY1821 pKa = 10.16SFLITNTGNVTLDD1834 pKa = 3.09RR1835 pKa = 11.84VAVDD1839 pKa = 3.19EE1840 pKa = 5.09GEE1842 pKa = 4.39FTGSGPLSPVTCPDD1856 pKa = 3.18EE1857 pKa = 5.22AGSLSPGEE1865 pKa = 4.15QATCTATYY1873 pKa = 9.45TVTQSDD1879 pKa = 3.65VDD1881 pKa = 3.87RR1882 pKa = 11.84GEE1884 pKa = 4.27LTNTATAGGTPPGGGGTTSPPSSATITGDD1913 pKa = 3.19AKK1915 pKa = 10.74PVLEE1919 pKa = 4.25VLKK1922 pKa = 9.21TAHH1925 pKa = 6.47WVDD1928 pKa = 4.12DD1929 pKa = 4.15NGSGVVDD1936 pKa = 4.64LGDD1939 pKa = 4.45RR1940 pKa = 11.84IAWTITATNTGGATATDD1957 pKa = 4.02VVVSDD1962 pKa = 4.06PTAGAVTCPATTLAPGEE1979 pKa = 4.64SMDD1982 pKa = 4.24CTAPDD1987 pKa = 3.69HH1988 pKa = 6.74VVDD1991 pKa = 4.37EE1992 pKa = 4.98ADD1994 pKa = 3.36VAAGEE1999 pKa = 4.34VANTAIATGRR2009 pKa = 11.84GPGGDD2014 pKa = 3.73PVTSPPASSTVEE2026 pKa = 3.82VGRR2029 pKa = 11.84PGTPPGPQPEE2039 pKa = 4.45EE2040 pKa = 3.8PGGPGDD2046 pKa = 3.98LATTGRR2052 pKa = 11.84DD2053 pKa = 3.22VGLLLALAGALLAFGGLLFSGARR2076 pKa = 11.84RR2077 pKa = 11.84RR2078 pKa = 11.84RR2079 pKa = 11.84RR2080 pKa = 11.84SS2081 pKa = 3.02
Molecular weight: 203.84 kDa
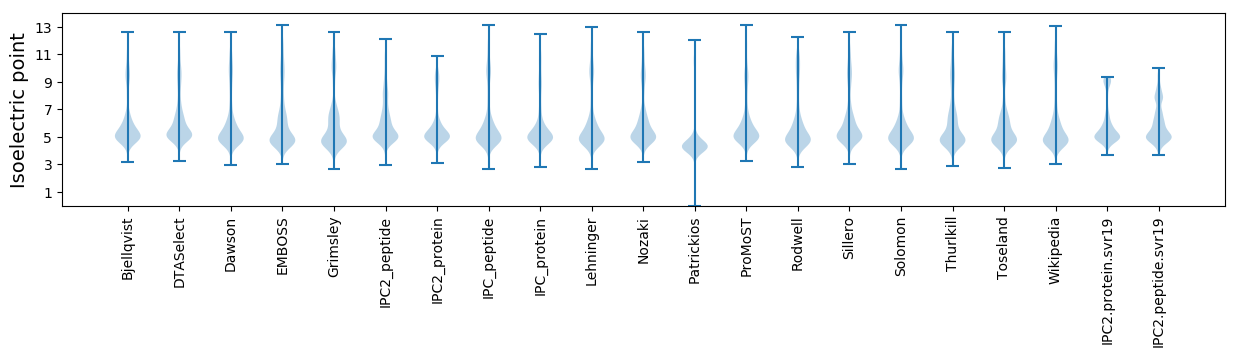
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6QSY8|A0A1I6QSY8_9PSEU PAS domain S-box-containing protein/diguanylate cyclase (GGDEF) domain-containing protein OS=Saccharopolyspora flava OX=95161 GN=SAMN05660874_01828 PE=4 SV=1
MM1 pKa = 7.27RR2 pKa = 11.84TLRR5 pKa = 11.84TGGALRR11 pKa = 11.84THH13 pKa = 5.08VTLRR17 pKa = 11.84AGGPLGSDD25 pKa = 3.49RR26 pKa = 11.84PLSTASPRR34 pKa = 11.84RR35 pKa = 11.84ALRR38 pKa = 11.84TGGPLGALSTLGALRR53 pKa = 11.84SRR55 pKa = 11.84RR56 pKa = 11.84SGQARR61 pKa = 11.84RR62 pKa = 11.84TLRR65 pKa = 11.84TGRR68 pKa = 11.84TLRR71 pKa = 11.84PRR73 pKa = 11.84GPLGARR79 pKa = 11.84SALSTRR85 pKa = 11.84SSLRR89 pKa = 11.84TSSALRR95 pKa = 11.84TRR97 pKa = 11.84CSLSTRR103 pKa = 11.84RR104 pKa = 11.84TLRR107 pKa = 11.84SGSALSTGRR116 pKa = 11.84TGSTLRR122 pKa = 11.84PRR124 pKa = 11.84RR125 pKa = 11.84TGRR128 pKa = 11.84TSRR131 pKa = 11.84ADD133 pKa = 3.3RR134 pKa = 11.84ALRR137 pKa = 11.84TRR139 pKa = 11.84GALSTGRR146 pKa = 11.84TRR148 pKa = 11.84STLRR152 pKa = 11.84TRR154 pKa = 11.84ITLRR158 pKa = 11.84PLSTRR163 pKa = 11.84STIRR167 pKa = 11.84TLRR170 pKa = 11.84ARR172 pKa = 11.84GTSRR176 pKa = 11.84TSGTDD181 pKa = 2.87LTLRR185 pKa = 11.84TRR187 pKa = 11.84SALRR191 pKa = 11.84TGLSLRR197 pKa = 11.84PRR199 pKa = 11.84RR200 pKa = 11.84TRR202 pKa = 11.84NAVSTLRR209 pKa = 11.84TRR211 pKa = 11.84RR212 pKa = 11.84ADD214 pKa = 3.12LTLRR218 pKa = 11.84TRR220 pKa = 11.84SALRR224 pKa = 11.84TRR226 pKa = 11.84RR227 pKa = 11.84TRR229 pKa = 11.84ITLRR233 pKa = 11.84PLRR236 pKa = 11.84TRR238 pKa = 11.84RR239 pKa = 11.84TGRR242 pKa = 11.84TSGTDD247 pKa = 2.96LTLRR251 pKa = 11.84TRR253 pKa = 11.84SALGTLRR260 pKa = 11.84TRR262 pKa = 11.84ISLRR266 pKa = 11.84ARR268 pKa = 11.84RR269 pKa = 11.84TRR271 pKa = 11.84STVSTLRR278 pKa = 11.84THH280 pKa = 7.21RR281 pKa = 11.84PDD283 pKa = 3.14LALRR287 pKa = 11.84PLRR290 pKa = 11.84TRR292 pKa = 11.84GAIGALRR299 pKa = 11.84THH301 pKa = 6.08RR302 pKa = 11.84TDD304 pKa = 2.88RR305 pKa = 11.84ALRR308 pKa = 11.84TRR310 pKa = 11.84RR311 pKa = 11.84TNITLRR317 pKa = 11.84PLRR320 pKa = 11.84SRR322 pKa = 11.84STVSSLRR329 pKa = 11.84TRR331 pKa = 11.84WADD334 pKa = 3.07LALRR338 pKa = 11.84TRR340 pKa = 11.84STGSALRR347 pKa = 11.84PRR349 pKa = 11.84GALSTIGTLRR359 pKa = 11.84THH361 pKa = 5.68RR362 pKa = 11.84TGRR365 pKa = 11.84PSRR368 pKa = 11.84TDD370 pKa = 2.95LALRR374 pKa = 11.84TRR376 pKa = 11.84SALRR380 pKa = 11.84TGRR383 pKa = 11.84ARR385 pKa = 11.84STLRR389 pKa = 11.84TRR391 pKa = 11.84ITLRR395 pKa = 11.84TLSTRR400 pKa = 11.84SAIGTLRR407 pKa = 11.84TRR409 pKa = 11.84RR410 pKa = 11.84TDD412 pKa = 3.02RR413 pKa = 11.84TSRR416 pKa = 11.84TDD418 pKa = 3.01LTLRR422 pKa = 11.84ARR424 pKa = 11.84STLRR428 pKa = 11.84TWGADD433 pKa = 3.06VALRR437 pKa = 11.84ALRR440 pKa = 11.84TGCAVGSGNALRR452 pKa = 11.84ADD454 pKa = 3.65GSLRR458 pKa = 11.84TGQPLWTGGALGTDD472 pKa = 3.41VALRR476 pKa = 11.84PLRR479 pKa = 11.84TGSARR484 pKa = 11.84RR485 pKa = 11.84TISALRR491 pKa = 11.84TNRR494 pKa = 11.84TRR496 pKa = 11.84RR497 pKa = 11.84ARR499 pKa = 11.84RR500 pKa = 11.84TDD502 pKa = 3.14LALRR506 pKa = 11.84SLNTGSARR514 pKa = 11.84GTVGTLRR521 pKa = 11.84TRR523 pKa = 11.84RR524 pKa = 11.84TIGALRR530 pKa = 11.84TGGTGRR536 pKa = 11.84ARR538 pKa = 11.84EE539 pKa = 4.06SLGTGVALRR548 pKa = 11.84PLRR551 pKa = 11.84ACGTRR556 pKa = 11.84RR557 pKa = 11.84AVGSLRR563 pKa = 11.84TGRR566 pKa = 11.84AHH568 pKa = 6.86RR569 pKa = 11.84AVGALRR575 pKa = 11.84ALRR578 pKa = 11.84SRR580 pKa = 11.84LARR583 pKa = 11.84ARR585 pKa = 11.84FSRR588 pKa = 11.84RR589 pKa = 11.84ALRR592 pKa = 11.84SFGSLGPGRR601 pKa = 11.84SGRR604 pKa = 11.84TRR606 pKa = 11.84DD607 pKa = 3.33AGSTGFALRR616 pKa = 11.84ANGPRR621 pKa = 11.84RR622 pKa = 11.84PGPALSTGGALRR634 pKa = 11.84AARR637 pKa = 11.84PSGWSLRR644 pKa = 11.84ALRR647 pKa = 11.84TVRR650 pKa = 11.84ALRR653 pKa = 11.84ALRR656 pKa = 11.84SFRR659 pKa = 11.84TGRR662 pKa = 11.84AGVRR666 pKa = 11.84SGRR669 pKa = 11.84QCRR672 pKa = 11.84RR673 pKa = 11.84GRR675 pKa = 11.84RR676 pKa = 11.84PRR678 pKa = 11.84LAEE681 pKa = 3.76HH682 pKa = 7.03RR683 pKa = 11.84SFHH686 pKa = 6.82PGRR689 pKa = 11.84THH691 pKa = 7.79AEE693 pKa = 3.75PGLFGTGDD701 pKa = 3.62GQRR704 pKa = 11.84GGRR707 pKa = 11.84SAQHH711 pKa = 6.34RR712 pKa = 11.84DD713 pKa = 3.12DD714 pKa = 4.1RR715 pKa = 11.84RR716 pKa = 11.84GGAHH720 pKa = 7.47DD721 pKa = 5.06DD722 pKa = 3.3VDD724 pKa = 3.85GAARR728 pKa = 11.84SSRR731 pKa = 11.84PATTHH736 pKa = 6.28EE737 pKa = 4.42SPPTILIVPLSVSCLDD753 pKa = 3.64SPIIGEE759 pKa = 4.38KK760 pKa = 8.92TSVWNPPFLNEE771 pKa = 4.05KK772 pKa = 10.2
MM1 pKa = 7.27RR2 pKa = 11.84TLRR5 pKa = 11.84TGGALRR11 pKa = 11.84THH13 pKa = 5.08VTLRR17 pKa = 11.84AGGPLGSDD25 pKa = 3.49RR26 pKa = 11.84PLSTASPRR34 pKa = 11.84RR35 pKa = 11.84ALRR38 pKa = 11.84TGGPLGALSTLGALRR53 pKa = 11.84SRR55 pKa = 11.84RR56 pKa = 11.84SGQARR61 pKa = 11.84RR62 pKa = 11.84TLRR65 pKa = 11.84TGRR68 pKa = 11.84TLRR71 pKa = 11.84PRR73 pKa = 11.84GPLGARR79 pKa = 11.84SALSTRR85 pKa = 11.84SSLRR89 pKa = 11.84TSSALRR95 pKa = 11.84TRR97 pKa = 11.84CSLSTRR103 pKa = 11.84RR104 pKa = 11.84TLRR107 pKa = 11.84SGSALSTGRR116 pKa = 11.84TGSTLRR122 pKa = 11.84PRR124 pKa = 11.84RR125 pKa = 11.84TGRR128 pKa = 11.84TSRR131 pKa = 11.84ADD133 pKa = 3.3RR134 pKa = 11.84ALRR137 pKa = 11.84TRR139 pKa = 11.84GALSTGRR146 pKa = 11.84TRR148 pKa = 11.84STLRR152 pKa = 11.84TRR154 pKa = 11.84ITLRR158 pKa = 11.84PLSTRR163 pKa = 11.84STIRR167 pKa = 11.84TLRR170 pKa = 11.84ARR172 pKa = 11.84GTSRR176 pKa = 11.84TSGTDD181 pKa = 2.87LTLRR185 pKa = 11.84TRR187 pKa = 11.84SALRR191 pKa = 11.84TGLSLRR197 pKa = 11.84PRR199 pKa = 11.84RR200 pKa = 11.84TRR202 pKa = 11.84NAVSTLRR209 pKa = 11.84TRR211 pKa = 11.84RR212 pKa = 11.84ADD214 pKa = 3.12LTLRR218 pKa = 11.84TRR220 pKa = 11.84SALRR224 pKa = 11.84TRR226 pKa = 11.84RR227 pKa = 11.84TRR229 pKa = 11.84ITLRR233 pKa = 11.84PLRR236 pKa = 11.84TRR238 pKa = 11.84RR239 pKa = 11.84TGRR242 pKa = 11.84TSGTDD247 pKa = 2.96LTLRR251 pKa = 11.84TRR253 pKa = 11.84SALGTLRR260 pKa = 11.84TRR262 pKa = 11.84ISLRR266 pKa = 11.84ARR268 pKa = 11.84RR269 pKa = 11.84TRR271 pKa = 11.84STVSTLRR278 pKa = 11.84THH280 pKa = 7.21RR281 pKa = 11.84PDD283 pKa = 3.14LALRR287 pKa = 11.84PLRR290 pKa = 11.84TRR292 pKa = 11.84GAIGALRR299 pKa = 11.84THH301 pKa = 6.08RR302 pKa = 11.84TDD304 pKa = 2.88RR305 pKa = 11.84ALRR308 pKa = 11.84TRR310 pKa = 11.84RR311 pKa = 11.84TNITLRR317 pKa = 11.84PLRR320 pKa = 11.84SRR322 pKa = 11.84STVSSLRR329 pKa = 11.84TRR331 pKa = 11.84WADD334 pKa = 3.07LALRR338 pKa = 11.84TRR340 pKa = 11.84STGSALRR347 pKa = 11.84PRR349 pKa = 11.84GALSTIGTLRR359 pKa = 11.84THH361 pKa = 5.68RR362 pKa = 11.84TGRR365 pKa = 11.84PSRR368 pKa = 11.84TDD370 pKa = 2.95LALRR374 pKa = 11.84TRR376 pKa = 11.84SALRR380 pKa = 11.84TGRR383 pKa = 11.84ARR385 pKa = 11.84STLRR389 pKa = 11.84TRR391 pKa = 11.84ITLRR395 pKa = 11.84TLSTRR400 pKa = 11.84SAIGTLRR407 pKa = 11.84TRR409 pKa = 11.84RR410 pKa = 11.84TDD412 pKa = 3.02RR413 pKa = 11.84TSRR416 pKa = 11.84TDD418 pKa = 3.01LTLRR422 pKa = 11.84ARR424 pKa = 11.84STLRR428 pKa = 11.84TWGADD433 pKa = 3.06VALRR437 pKa = 11.84ALRR440 pKa = 11.84TGCAVGSGNALRR452 pKa = 11.84ADD454 pKa = 3.65GSLRR458 pKa = 11.84TGQPLWTGGALGTDD472 pKa = 3.41VALRR476 pKa = 11.84PLRR479 pKa = 11.84TGSARR484 pKa = 11.84RR485 pKa = 11.84TISALRR491 pKa = 11.84TNRR494 pKa = 11.84TRR496 pKa = 11.84RR497 pKa = 11.84ARR499 pKa = 11.84RR500 pKa = 11.84TDD502 pKa = 3.14LALRR506 pKa = 11.84SLNTGSARR514 pKa = 11.84GTVGTLRR521 pKa = 11.84TRR523 pKa = 11.84RR524 pKa = 11.84TIGALRR530 pKa = 11.84TGGTGRR536 pKa = 11.84ARR538 pKa = 11.84EE539 pKa = 4.06SLGTGVALRR548 pKa = 11.84PLRR551 pKa = 11.84ACGTRR556 pKa = 11.84RR557 pKa = 11.84AVGSLRR563 pKa = 11.84TGRR566 pKa = 11.84AHH568 pKa = 6.86RR569 pKa = 11.84AVGALRR575 pKa = 11.84ALRR578 pKa = 11.84SRR580 pKa = 11.84LARR583 pKa = 11.84ARR585 pKa = 11.84FSRR588 pKa = 11.84RR589 pKa = 11.84ALRR592 pKa = 11.84SFGSLGPGRR601 pKa = 11.84SGRR604 pKa = 11.84TRR606 pKa = 11.84DD607 pKa = 3.33AGSTGFALRR616 pKa = 11.84ANGPRR621 pKa = 11.84RR622 pKa = 11.84PGPALSTGGALRR634 pKa = 11.84AARR637 pKa = 11.84PSGWSLRR644 pKa = 11.84ALRR647 pKa = 11.84TVRR650 pKa = 11.84ALRR653 pKa = 11.84ALRR656 pKa = 11.84SFRR659 pKa = 11.84TGRR662 pKa = 11.84AGVRR666 pKa = 11.84SGRR669 pKa = 11.84QCRR672 pKa = 11.84RR673 pKa = 11.84GRR675 pKa = 11.84RR676 pKa = 11.84PRR678 pKa = 11.84LAEE681 pKa = 3.76HH682 pKa = 7.03RR683 pKa = 11.84SFHH686 pKa = 6.82PGRR689 pKa = 11.84THH691 pKa = 7.79AEE693 pKa = 3.75PGLFGTGDD701 pKa = 3.62GQRR704 pKa = 11.84GGRR707 pKa = 11.84SAQHH711 pKa = 6.34RR712 pKa = 11.84DD713 pKa = 3.12DD714 pKa = 4.1RR715 pKa = 11.84RR716 pKa = 11.84GGAHH720 pKa = 7.47DD721 pKa = 5.06DD722 pKa = 3.3VDD724 pKa = 3.85GAARR728 pKa = 11.84SSRR731 pKa = 11.84PATTHH736 pKa = 6.28EE737 pKa = 4.42SPPTILIVPLSVSCLDD753 pKa = 3.64SPIIGEE759 pKa = 4.38KK760 pKa = 8.92TSVWNPPFLNEE771 pKa = 4.05KK772 pKa = 10.2
Molecular weight: 84.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1855314 |
39 |
10081 |
331.6 |
35.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.732 ± 0.051 | 0.787 ± 0.01 |
6.223 ± 0.029 | 6.236 ± 0.032 |
2.933 ± 0.02 | 9.195 ± 0.037 |
2.316 ± 0.017 | 3.511 ± 0.024 |
2.016 ± 0.028 | 10.379 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.839 ± 0.016 | 1.936 ± 0.018 |
5.928 ± 0.034 | 3.023 ± 0.023 |
7.819 ± 0.04 | 5.283 ± 0.032 |
5.788 ± 0.032 | 8.723 ± 0.034 |
1.467 ± 0.013 | 1.866 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
