
Sesbania mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
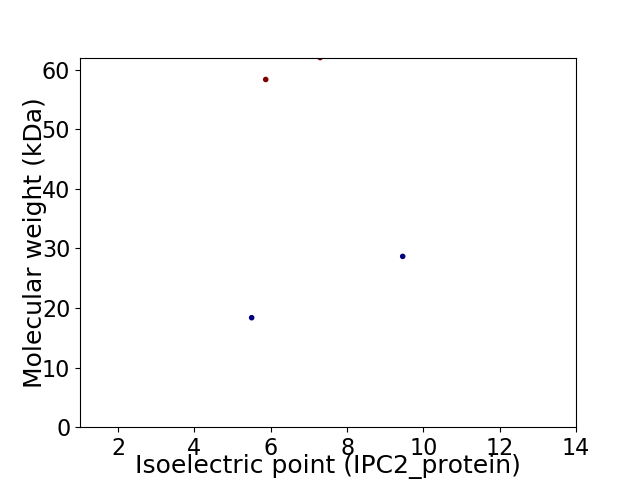
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9EB09|Q9EB09_9VIRU Putative movement protein OS=Sesbania mosaic virus OX=12558 PE=4 SV=1
MM1 pKa = 7.06QAQHH5 pKa = 6.06TFTIKK10 pKa = 10.58FLRR13 pKa = 11.84HH14 pKa = 4.65ACFIGFEE21 pKa = 4.24DD22 pKa = 3.78PRR24 pKa = 11.84VVLDD28 pKa = 4.12HH29 pKa = 6.96EE30 pKa = 5.35EE31 pKa = 3.66IDD33 pKa = 3.71IPCEE37 pKa = 4.19VEE39 pKa = 3.77FDD41 pKa = 4.2CNAEE45 pKa = 4.35SVCLVRR51 pKa = 11.84AHH53 pKa = 5.79NQPYY57 pKa = 10.39SEE59 pKa = 4.8GEE61 pKa = 4.07VKK63 pKa = 10.66NYY65 pKa = 10.35SVYY68 pKa = 10.54FNSGLNDD75 pKa = 3.38YY76 pKa = 10.0YY77 pKa = 11.57GPLLPVWLEE86 pKa = 3.7IVCRR90 pKa = 11.84VCATSYY96 pKa = 11.25SFVLAPEE103 pKa = 4.45DD104 pKa = 4.28LEE106 pKa = 6.03DD107 pKa = 3.44EE108 pKa = 4.58AGRR111 pKa = 11.84VSRR114 pKa = 11.84KK115 pKa = 8.96YY116 pKa = 10.72APCEE120 pKa = 3.67NGKK123 pKa = 9.37VFQRR127 pKa = 11.84AVRR130 pKa = 11.84RR131 pKa = 11.84ATGEE135 pKa = 3.93EE136 pKa = 4.18YY137 pKa = 10.27FYY139 pKa = 11.48SCRR142 pKa = 11.84KK143 pKa = 8.72KK144 pKa = 10.39VCRR147 pKa = 11.84EE148 pKa = 3.93CIIRR152 pKa = 11.84AARR155 pKa = 11.84AMSSSS160 pKa = 3.21
MM1 pKa = 7.06QAQHH5 pKa = 6.06TFTIKK10 pKa = 10.58FLRR13 pKa = 11.84HH14 pKa = 4.65ACFIGFEE21 pKa = 4.24DD22 pKa = 3.78PRR24 pKa = 11.84VVLDD28 pKa = 4.12HH29 pKa = 6.96EE30 pKa = 5.35EE31 pKa = 3.66IDD33 pKa = 3.71IPCEE37 pKa = 4.19VEE39 pKa = 3.77FDD41 pKa = 4.2CNAEE45 pKa = 4.35SVCLVRR51 pKa = 11.84AHH53 pKa = 5.79NQPYY57 pKa = 10.39SEE59 pKa = 4.8GEE61 pKa = 4.07VKK63 pKa = 10.66NYY65 pKa = 10.35SVYY68 pKa = 10.54FNSGLNDD75 pKa = 3.38YY76 pKa = 10.0YY77 pKa = 11.57GPLLPVWLEE86 pKa = 3.7IVCRR90 pKa = 11.84VCATSYY96 pKa = 11.25SFVLAPEE103 pKa = 4.45DD104 pKa = 4.28LEE106 pKa = 6.03DD107 pKa = 3.44EE108 pKa = 4.58AGRR111 pKa = 11.84VSRR114 pKa = 11.84KK115 pKa = 8.96YY116 pKa = 10.72APCEE120 pKa = 3.67NGKK123 pKa = 9.37VFQRR127 pKa = 11.84AVRR130 pKa = 11.84RR131 pKa = 11.84ATGEE135 pKa = 3.93EE136 pKa = 4.18YY137 pKa = 10.27FYY139 pKa = 11.48SCRR142 pKa = 11.84KK143 pKa = 8.72KK144 pKa = 10.39VCRR147 pKa = 11.84EE148 pKa = 3.93CIIRR152 pKa = 11.84AARR155 pKa = 11.84AMSSSS160 pKa = 3.21
Molecular weight: 18.37 kDa
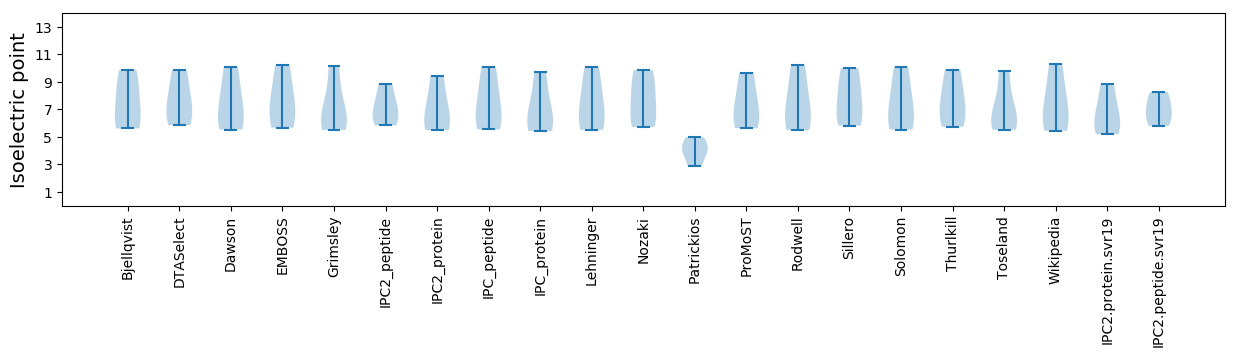
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9EB07|Q9EB07_9VIRU RNA-directed RNA polymerase OS=Sesbania mosaic virus OX=12558 PE=4 SV=2
MM1 pKa = 7.88AKK3 pKa = 10.14RR4 pKa = 11.84LSKK7 pKa = 10.44QQLAKK12 pKa = 10.73AIANTLEE19 pKa = 4.36TPPQPKK25 pKa = 9.71AGRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84NRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84QRR36 pKa = 11.84SAVQQLQPTQAGISMAPSAQGAMVRR61 pKa = 11.84IRR63 pKa = 11.84NPAVSSSRR71 pKa = 11.84GGITVLTHH79 pKa = 6.31SEE81 pKa = 3.87LSAEE85 pKa = 4.11IGVTDD90 pKa = 4.41SIVVSSEE97 pKa = 3.28LVMPYY102 pKa = 9.63TVGTWLRR109 pKa = 11.84GVAANWSKK117 pKa = 11.18YY118 pKa = 7.01SWLSVRR124 pKa = 11.84YY125 pKa = 7.63TYY127 pKa = 10.52IPSCPSSTAGSIHH140 pKa = 6.68MGFQYY145 pKa = 11.56DD146 pKa = 3.5MADD149 pKa = 3.54TVPVSVNQLSNLRR162 pKa = 11.84GYY164 pKa = 11.09VSGQVWSGSAGLCFINGTRR183 pKa = 11.84CSDD186 pKa = 3.0TSTAISTTLDD196 pKa = 3.19VSKK199 pKa = 10.62LGKK202 pKa = 9.35KK203 pKa = 8.4WYY205 pKa = 8.96PYY207 pKa = 8.6KK208 pKa = 10.61TSADD212 pKa = 3.43YY213 pKa = 8.57ATAVGVDD220 pKa = 3.59VNIATPLVPARR231 pKa = 11.84LVIALLDD238 pKa = 4.31GSSSTAVAAGRR249 pKa = 11.84IYY251 pKa = 10.13CTYY254 pKa = 8.53TIQMIEE260 pKa = 4.11PTASALNNN268 pKa = 3.59
MM1 pKa = 7.88AKK3 pKa = 10.14RR4 pKa = 11.84LSKK7 pKa = 10.44QQLAKK12 pKa = 10.73AIANTLEE19 pKa = 4.36TPPQPKK25 pKa = 9.71AGRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84NRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84QRR36 pKa = 11.84SAVQQLQPTQAGISMAPSAQGAMVRR61 pKa = 11.84IRR63 pKa = 11.84NPAVSSSRR71 pKa = 11.84GGITVLTHH79 pKa = 6.31SEE81 pKa = 3.87LSAEE85 pKa = 4.11IGVTDD90 pKa = 4.41SIVVSSEE97 pKa = 3.28LVMPYY102 pKa = 9.63TVGTWLRR109 pKa = 11.84GVAANWSKK117 pKa = 11.18YY118 pKa = 7.01SWLSVRR124 pKa = 11.84YY125 pKa = 7.63TYY127 pKa = 10.52IPSCPSSTAGSIHH140 pKa = 6.68MGFQYY145 pKa = 11.56DD146 pKa = 3.5MADD149 pKa = 3.54TVPVSVNQLSNLRR162 pKa = 11.84GYY164 pKa = 11.09VSGQVWSGSAGLCFINGTRR183 pKa = 11.84CSDD186 pKa = 3.0TSTAISTTLDD196 pKa = 3.19VSKK199 pKa = 10.62LGKK202 pKa = 9.35KK203 pKa = 8.4WYY205 pKa = 8.96PYY207 pKa = 8.6KK208 pKa = 10.61TSADD212 pKa = 3.43YY213 pKa = 8.57ATAVGVDD220 pKa = 3.59VNIATPLVPARR231 pKa = 11.84LVIALLDD238 pKa = 4.31GSSSTAVAAGRR249 pKa = 11.84IYY251 pKa = 10.13CTYY254 pKa = 8.53TIQMIEE260 pKa = 4.11PTASALNNN268 pKa = 3.59
Molecular weight: 28.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1521 |
160 |
572 |
380.3 |
41.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.284 ± 0.654 | 3.287 ± 0.715 |
3.813 ± 0.356 | 6.377 ± 1.233 |
3.024 ± 0.771 | 6.443 ± 0.343 |
1.775 ± 0.267 | 3.813 ± 0.479 |
5.128 ± 0.633 | 9.007 ± 0.839 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.104 ± 0.236 | 2.893 ± 0.297 |
6.772 ± 0.75 | 3.287 ± 0.371 |
5.588 ± 0.889 | 10.717 ± 0.942 |
5.194 ± 0.981 | 7.101 ± 0.678 |
1.972 ± 0.303 | 3.419 ± 0.448 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
