
Wenling crustacean virus 10
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alphacrustrhavirus; Wenling alphacrustrhavirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
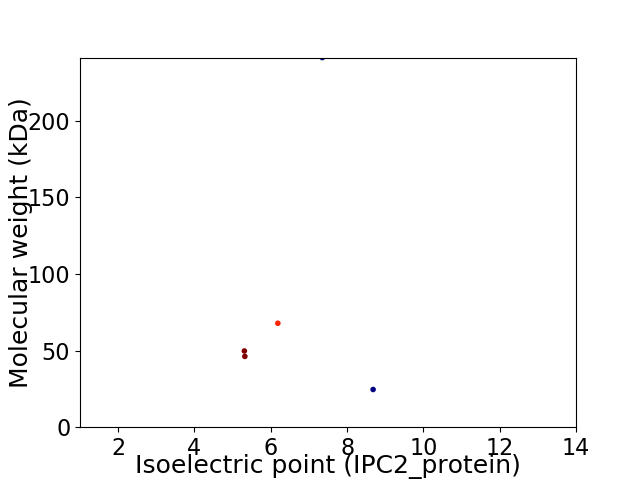
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
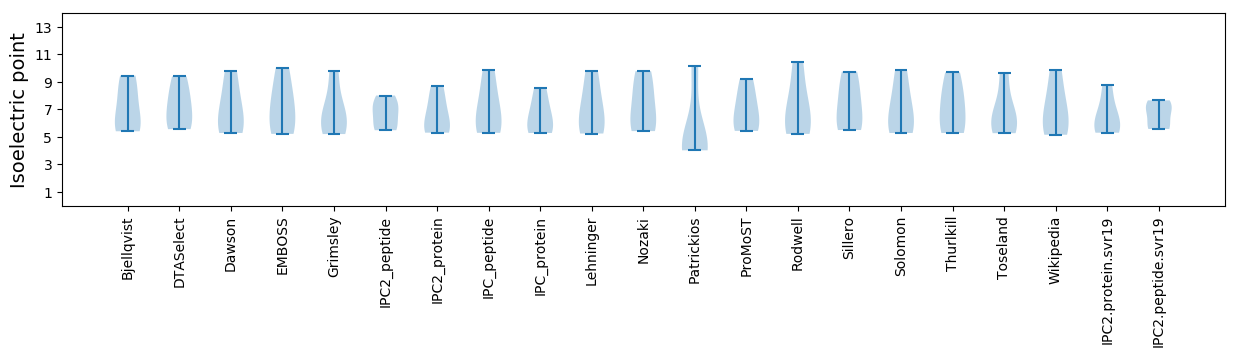
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNA1|A0A1L3KNA1_9VIRU Uncharacterized protein OS=Wenling crustacean virus 10 OX=1923479 PE=4 SV=1
MM1 pKa = 6.95TRR3 pKa = 11.84KK4 pKa = 10.21KK5 pKa = 10.49IQDD8 pKa = 4.05PCNMADD14 pKa = 3.59PAYY17 pKa = 9.9QALSGLPSRR26 pKa = 11.84LVDD29 pKa = 3.67VQDD32 pKa = 4.82KK33 pKa = 10.59GWSDD37 pKa = 4.3DD38 pKa = 3.71SLKK41 pKa = 10.82KK42 pKa = 10.44YY43 pKa = 10.15IKK45 pKa = 10.07SRR47 pKa = 11.84YY48 pKa = 8.42SLPGEE53 pKa = 4.23DD54 pKa = 6.09DD55 pKa = 5.33RR56 pKa = 11.84ITLCKK61 pKa = 10.44LFLQAYY67 pKa = 10.02CYY69 pKa = 11.21GNWQNFKK76 pKa = 10.64LSEE79 pKa = 4.1IVHH82 pKa = 6.61ALLLITPTRR91 pKa = 11.84TEE93 pKa = 3.82KK94 pKa = 10.73KK95 pKa = 9.56YY96 pKa = 10.75LALFACFKK104 pKa = 9.88TKK106 pKa = 10.22QSPLIDD112 pKa = 3.62LSGAKK117 pKa = 9.17VQPADD122 pKa = 4.03DD123 pKa = 4.27ATVSDD128 pKa = 4.84PKK130 pKa = 10.61TSQKK134 pKa = 11.34DD135 pKa = 3.54DD136 pKa = 3.78TNDD139 pKa = 3.31LQGLVGEE146 pKa = 5.18HH147 pKa = 6.96DD148 pKa = 4.6WIDD151 pKa = 3.91DD152 pKa = 3.62DD153 pKa = 5.93DD154 pKa = 6.23NIVTGLPRR162 pKa = 11.84NTSSFYY168 pKa = 11.06KK169 pKa = 10.85GMLEE173 pKa = 5.36ALDD176 pKa = 4.58GDD178 pKa = 4.46DD179 pKa = 4.84KK180 pKa = 11.12IAAIRR185 pKa = 11.84FCCFLAWVCFRR196 pKa = 11.84TVVKK200 pKa = 10.2EE201 pKa = 4.11VSSVRR206 pKa = 11.84AYY208 pKa = 10.82LGRR211 pKa = 11.84AEE213 pKa = 4.1NFSRR217 pKa = 11.84LVTNLTDD224 pKa = 3.97FEE226 pKa = 4.74LPEE229 pKa = 4.35KK230 pKa = 10.94VPLPSKK236 pKa = 10.84IMLEE240 pKa = 4.27KK241 pKa = 10.33IAITMGKK248 pKa = 7.83GTASVRR254 pKa = 11.84EE255 pKa = 4.28LISWAVYY262 pKa = 10.27VKK264 pKa = 10.34VHH266 pKa = 6.74SDD268 pKa = 3.07NDD270 pKa = 3.62DD271 pKa = 3.62DD272 pKa = 4.17VRR274 pKa = 11.84MMEE277 pKa = 4.69GGCLTHH283 pKa = 8.11LSEE286 pKa = 4.94NGIGLVGLVDD296 pKa = 3.73KK297 pKa = 11.1VAGVFSVSIEE307 pKa = 4.33EE308 pKa = 4.05ILSLILTSHH317 pKa = 6.13TMNSVIRR324 pKa = 11.84LADD327 pKa = 4.71FIKK330 pKa = 10.7HH331 pKa = 4.98VVPKK335 pKa = 9.96VKK337 pKa = 9.13SWKK340 pKa = 7.53WSRR343 pKa = 11.84VIDD346 pKa = 4.07DD347 pKa = 3.63QCFSSLKK354 pKa = 10.28ILDD357 pKa = 4.62NIVLSAYY364 pKa = 10.19LVALVTEE371 pKa = 4.78ADD373 pKa = 3.98AGNDD377 pKa = 3.08MWNMVVFEE385 pKa = 3.96QHH387 pKa = 6.64LPNTTKK393 pKa = 10.76NQALRR398 pKa = 11.84WAKK401 pKa = 10.06KK402 pKa = 8.87YY403 pKa = 10.16VVASSEE409 pKa = 4.1HH410 pKa = 5.58DD411 pKa = 3.35QKK413 pKa = 10.17TGAWVSEE420 pKa = 3.93KK421 pKa = 10.23HH422 pKa = 5.52RR423 pKa = 11.84KK424 pKa = 7.27IWAYY428 pKa = 10.63GEE430 pKa = 5.11DD431 pKa = 4.43DD432 pKa = 4.51DD433 pKa = 6.41EE434 pKa = 6.93SDD436 pKa = 3.63NEE438 pKa = 4.12YY439 pKa = 11.16SGLII443 pKa = 3.74
MM1 pKa = 6.95TRR3 pKa = 11.84KK4 pKa = 10.21KK5 pKa = 10.49IQDD8 pKa = 4.05PCNMADD14 pKa = 3.59PAYY17 pKa = 9.9QALSGLPSRR26 pKa = 11.84LVDD29 pKa = 3.67VQDD32 pKa = 4.82KK33 pKa = 10.59GWSDD37 pKa = 4.3DD38 pKa = 3.71SLKK41 pKa = 10.82KK42 pKa = 10.44YY43 pKa = 10.15IKK45 pKa = 10.07SRR47 pKa = 11.84YY48 pKa = 8.42SLPGEE53 pKa = 4.23DD54 pKa = 6.09DD55 pKa = 5.33RR56 pKa = 11.84ITLCKK61 pKa = 10.44LFLQAYY67 pKa = 10.02CYY69 pKa = 11.21GNWQNFKK76 pKa = 10.64LSEE79 pKa = 4.1IVHH82 pKa = 6.61ALLLITPTRR91 pKa = 11.84TEE93 pKa = 3.82KK94 pKa = 10.73KK95 pKa = 9.56YY96 pKa = 10.75LALFACFKK104 pKa = 9.88TKK106 pKa = 10.22QSPLIDD112 pKa = 3.62LSGAKK117 pKa = 9.17VQPADD122 pKa = 4.03DD123 pKa = 4.27ATVSDD128 pKa = 4.84PKK130 pKa = 10.61TSQKK134 pKa = 11.34DD135 pKa = 3.54DD136 pKa = 3.78TNDD139 pKa = 3.31LQGLVGEE146 pKa = 5.18HH147 pKa = 6.96DD148 pKa = 4.6WIDD151 pKa = 3.91DD152 pKa = 3.62DD153 pKa = 5.93DD154 pKa = 6.23NIVTGLPRR162 pKa = 11.84NTSSFYY168 pKa = 11.06KK169 pKa = 10.85GMLEE173 pKa = 5.36ALDD176 pKa = 4.58GDD178 pKa = 4.46DD179 pKa = 4.84KK180 pKa = 11.12IAAIRR185 pKa = 11.84FCCFLAWVCFRR196 pKa = 11.84TVVKK200 pKa = 10.2EE201 pKa = 4.11VSSVRR206 pKa = 11.84AYY208 pKa = 10.82LGRR211 pKa = 11.84AEE213 pKa = 4.1NFSRR217 pKa = 11.84LVTNLTDD224 pKa = 3.97FEE226 pKa = 4.74LPEE229 pKa = 4.35KK230 pKa = 10.94VPLPSKK236 pKa = 10.84IMLEE240 pKa = 4.27KK241 pKa = 10.33IAITMGKK248 pKa = 7.83GTASVRR254 pKa = 11.84EE255 pKa = 4.28LISWAVYY262 pKa = 10.27VKK264 pKa = 10.34VHH266 pKa = 6.74SDD268 pKa = 3.07NDD270 pKa = 3.62DD271 pKa = 3.62DD272 pKa = 4.17VRR274 pKa = 11.84MMEE277 pKa = 4.69GGCLTHH283 pKa = 8.11LSEE286 pKa = 4.94NGIGLVGLVDD296 pKa = 3.73KK297 pKa = 11.1VAGVFSVSIEE307 pKa = 4.33EE308 pKa = 4.05ILSLILTSHH317 pKa = 6.13TMNSVIRR324 pKa = 11.84LADD327 pKa = 4.71FIKK330 pKa = 10.7HH331 pKa = 4.98VVPKK335 pKa = 9.96VKK337 pKa = 9.13SWKK340 pKa = 7.53WSRR343 pKa = 11.84VIDD346 pKa = 4.07DD347 pKa = 3.63QCFSSLKK354 pKa = 10.28ILDD357 pKa = 4.62NIVLSAYY364 pKa = 10.19LVALVTEE371 pKa = 4.78ADD373 pKa = 3.98AGNDD377 pKa = 3.08MWNMVVFEE385 pKa = 3.96QHH387 pKa = 6.64LPNTTKK393 pKa = 10.76NQALRR398 pKa = 11.84WAKK401 pKa = 10.06KK402 pKa = 8.87YY403 pKa = 10.16VVASSEE409 pKa = 4.1HH410 pKa = 5.58DD411 pKa = 3.35QKK413 pKa = 10.17TGAWVSEE420 pKa = 3.93KK421 pKa = 10.23HH422 pKa = 5.52RR423 pKa = 11.84KK424 pKa = 7.27IWAYY428 pKa = 10.63GEE430 pKa = 5.11DD431 pKa = 4.43DD432 pKa = 4.51DD433 pKa = 6.41EE434 pKa = 6.93SDD436 pKa = 3.63NEE438 pKa = 4.12YY439 pKa = 11.16SGLII443 pKa = 3.74
Molecular weight: 49.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KN39|A0A1L3KN39_9VIRU Uncharacterized protein OS=Wenling crustacean virus 10 OX=1923479 PE=4 SV=1
MM1 pKa = 6.96QCRR4 pKa = 11.84FRR6 pKa = 11.84KK7 pKa = 10.08NKK9 pKa = 9.23TPEE12 pKa = 4.0KK13 pKa = 10.26LDD15 pKa = 3.16IMSNLVIKK23 pKa = 10.52LCAEE27 pKa = 3.98GTIVGTPAHH36 pKa = 6.5SWAQIASTIVDD47 pKa = 4.36ALFKK51 pKa = 11.06DD52 pKa = 4.38CGFSPKK58 pKa = 9.64IMAQIIVLLHH68 pKa = 5.85EE69 pKa = 4.39SLRR72 pKa = 11.84DD73 pKa = 3.69KK74 pKa = 11.1VQEE77 pKa = 4.1TPVRR81 pKa = 11.84RR82 pKa = 11.84TIISLKK88 pKa = 10.94DD89 pKa = 3.59EE90 pKa = 4.5IEE92 pKa = 4.33KK93 pKa = 10.64EE94 pKa = 4.25WVTKK98 pKa = 9.35RR99 pKa = 11.84TYY101 pKa = 9.6NCKK104 pKa = 10.03LVAHH108 pKa = 6.56TSFPYY113 pKa = 10.73SPDD116 pKa = 3.18LNGFLKK122 pKa = 10.65KK123 pKa = 10.69LNLEE127 pKa = 4.06LSGEE131 pKa = 4.34GKK133 pKa = 10.39EE134 pKa = 4.32GTVVVSLRR142 pKa = 11.84LQGEE146 pKa = 4.45LRR148 pKa = 11.84CKK150 pKa = 8.18PTNYY154 pKa = 10.11DD155 pKa = 3.21PQTIQDD161 pKa = 4.0LRR163 pKa = 11.84FVPLPPVGKK172 pKa = 10.39VLTEE176 pKa = 4.0QISKK180 pKa = 10.43LRR182 pKa = 11.84AKK184 pKa = 9.26TKK186 pKa = 10.62NQIEE190 pKa = 4.4VPKK193 pKa = 10.87DD194 pKa = 2.99HH195 pKa = 6.73GQDD198 pKa = 3.24EE199 pKa = 5.05GKK201 pKa = 10.51GASSKK206 pKa = 10.84GRR208 pKa = 11.84LKK210 pKa = 11.21GLFSGLKK217 pKa = 10.12AKK219 pKa = 9.98EE220 pKa = 3.77
MM1 pKa = 6.96QCRR4 pKa = 11.84FRR6 pKa = 11.84KK7 pKa = 10.08NKK9 pKa = 9.23TPEE12 pKa = 4.0KK13 pKa = 10.26LDD15 pKa = 3.16IMSNLVIKK23 pKa = 10.52LCAEE27 pKa = 3.98GTIVGTPAHH36 pKa = 6.5SWAQIASTIVDD47 pKa = 4.36ALFKK51 pKa = 11.06DD52 pKa = 4.38CGFSPKK58 pKa = 9.64IMAQIIVLLHH68 pKa = 5.85EE69 pKa = 4.39SLRR72 pKa = 11.84DD73 pKa = 3.69KK74 pKa = 11.1VQEE77 pKa = 4.1TPVRR81 pKa = 11.84RR82 pKa = 11.84TIISLKK88 pKa = 10.94DD89 pKa = 3.59EE90 pKa = 4.5IEE92 pKa = 4.33KK93 pKa = 10.64EE94 pKa = 4.25WVTKK98 pKa = 9.35RR99 pKa = 11.84TYY101 pKa = 9.6NCKK104 pKa = 10.03LVAHH108 pKa = 6.56TSFPYY113 pKa = 10.73SPDD116 pKa = 3.18LNGFLKK122 pKa = 10.65KK123 pKa = 10.69LNLEE127 pKa = 4.06LSGEE131 pKa = 4.34GKK133 pKa = 10.39EE134 pKa = 4.32GTVVVSLRR142 pKa = 11.84LQGEE146 pKa = 4.45LRR148 pKa = 11.84CKK150 pKa = 8.18PTNYY154 pKa = 10.11DD155 pKa = 3.21PQTIQDD161 pKa = 4.0LRR163 pKa = 11.84FVPLPPVGKK172 pKa = 10.39VLTEE176 pKa = 4.0QISKK180 pKa = 10.43LRR182 pKa = 11.84AKK184 pKa = 9.26TKK186 pKa = 10.62NQIEE190 pKa = 4.4VPKK193 pKa = 10.87DD194 pKa = 2.99HH195 pKa = 6.73GQDD198 pKa = 3.24EE199 pKa = 5.05GKK201 pKa = 10.51GASSKK206 pKa = 10.84GRR208 pKa = 11.84LKK210 pKa = 11.21GLFSGLKK217 pKa = 10.12AKK219 pKa = 9.98EE220 pKa = 3.77
Molecular weight: 24.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3799 |
220 |
2121 |
759.8 |
85.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.764 ± 0.512 | 1.764 ± 0.296 |
5.212 ± 0.848 | 6.554 ± 0.822 |
3.948 ± 0.314 | 5.791 ± 0.21 |
2.316 ± 0.18 | 6.002 ± 0.552 |
7.265 ± 1.296 | 11.345 ± 0.46 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.343 | 3.343 ± 0.203 |
4.606 ± 0.302 | 3.948 ± 0.278 |
4.396 ± 0.555 | 8.95 ± 0.383 |
6.712 ± 0.552 | 5.844 ± 0.429 |
2.211 ± 0.326 | 2.843 ± 0.229 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
