
Sphingomonas sp. FARSPH
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
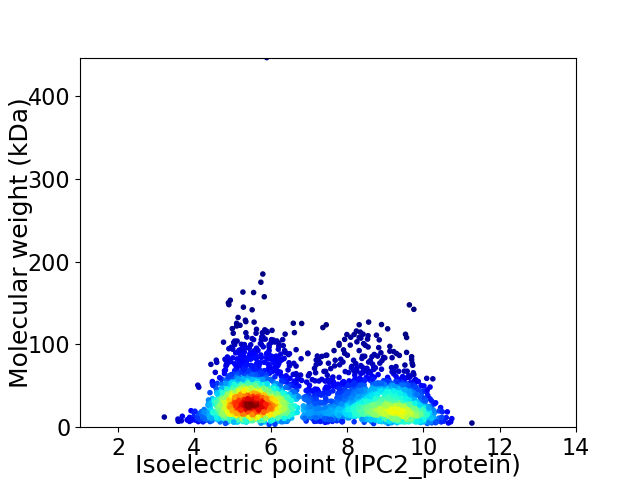
Virtual 2D-PAGE plot for 3408 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345WLE7|A0A345WLE7_9SPHN CopG family transcriptional regulator OS=Sphingomonas sp. FARSPH OX=2219696 GN=DM480_02345 PE=4 SV=1
MM1 pKa = 7.41TDD3 pKa = 3.15AAEE6 pKa = 4.37SNPRR10 pKa = 11.84IYY12 pKa = 10.29VACLAAYY19 pKa = 10.35NNGYY23 pKa = 9.03LHH25 pKa = 6.64GAWIDD30 pKa = 3.46ADD32 pKa = 3.48QDD34 pKa = 3.29ADD36 pKa = 4.25EE37 pKa = 4.78IRR39 pKa = 11.84SEE41 pKa = 3.88IAAMLSRR48 pKa = 11.84SPIEE52 pKa = 4.05GAEE55 pKa = 4.02EE56 pKa = 4.06YY57 pKa = 10.53AIHH60 pKa = 7.57DD61 pKa = 3.89YY62 pKa = 11.51EE63 pKa = 4.61GFEE66 pKa = 4.05GVAIEE71 pKa = 4.19EE72 pKa = 4.18FAGIEE77 pKa = 3.95NVARR81 pKa = 11.84IAAFVAEE88 pKa = 4.86HH89 pKa = 6.38GALGAGLLNEE99 pKa = 4.36FVGDD103 pKa = 3.38IDD105 pKa = 3.87QAEE108 pKa = 4.2TAMRR112 pKa = 11.84DD113 pKa = 3.77CYY115 pKa = 10.59HH116 pKa = 6.39GQFTSLADD124 pKa = 3.6YY125 pKa = 10.44MEE127 pKa = 5.11EE128 pKa = 4.0LTADD132 pKa = 3.53TVTIPDD138 pKa = 3.66ALRR141 pKa = 11.84YY142 pKa = 9.57YY143 pKa = 10.25IDD145 pKa = 3.54WQAMARR151 pKa = 11.84DD152 pKa = 3.99AEE154 pKa = 4.37LGGDD158 pKa = 4.28LFTVEE163 pKa = 3.76TAYY166 pKa = 10.96DD167 pKa = 3.77EE168 pKa = 4.3VHH170 pKa = 5.62VFSRR174 pKa = 11.84RR175 pKa = 3.3
MM1 pKa = 7.41TDD3 pKa = 3.15AAEE6 pKa = 4.37SNPRR10 pKa = 11.84IYY12 pKa = 10.29VACLAAYY19 pKa = 10.35NNGYY23 pKa = 9.03LHH25 pKa = 6.64GAWIDD30 pKa = 3.46ADD32 pKa = 3.48QDD34 pKa = 3.29ADD36 pKa = 4.25EE37 pKa = 4.78IRR39 pKa = 11.84SEE41 pKa = 3.88IAAMLSRR48 pKa = 11.84SPIEE52 pKa = 4.05GAEE55 pKa = 4.02EE56 pKa = 4.06YY57 pKa = 10.53AIHH60 pKa = 7.57DD61 pKa = 3.89YY62 pKa = 11.51EE63 pKa = 4.61GFEE66 pKa = 4.05GVAIEE71 pKa = 4.19EE72 pKa = 4.18FAGIEE77 pKa = 3.95NVARR81 pKa = 11.84IAAFVAEE88 pKa = 4.86HH89 pKa = 6.38GALGAGLLNEE99 pKa = 4.36FVGDD103 pKa = 3.38IDD105 pKa = 3.87QAEE108 pKa = 4.2TAMRR112 pKa = 11.84DD113 pKa = 3.77CYY115 pKa = 10.59HH116 pKa = 6.39GQFTSLADD124 pKa = 3.6YY125 pKa = 10.44MEE127 pKa = 5.11EE128 pKa = 4.0LTADD132 pKa = 3.53TVTIPDD138 pKa = 3.66ALRR141 pKa = 11.84YY142 pKa = 9.57YY143 pKa = 10.25IDD145 pKa = 3.54WQAMARR151 pKa = 11.84DD152 pKa = 3.99AEE154 pKa = 4.37LGGDD158 pKa = 4.28LFTVEE163 pKa = 3.76TAYY166 pKa = 10.96DD167 pKa = 3.77EE168 pKa = 4.3VHH170 pKa = 5.62VFSRR174 pKa = 11.84RR175 pKa = 3.3
Molecular weight: 19.3 kDa
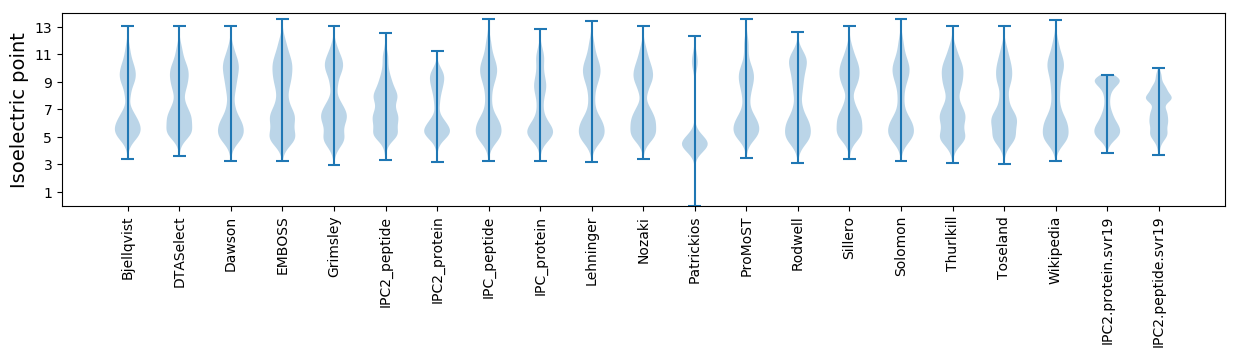
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345WT58|A0A345WT58_9SPHN Putative pterin-4-alpha-carbinolamine dehydratase OS=Sphingomonas sp. FARSPH OX=2219696 GN=DM480_05330 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1101842 |
29 |
4693 |
323.3 |
34.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.595 ± 0.079 | 0.709 ± 0.012 |
6.145 ± 0.036 | 4.605 ± 0.044 |
3.325 ± 0.021 | 9.078 ± 0.047 |
1.967 ± 0.024 | 4.739 ± 0.028 |
2.537 ± 0.033 | 9.784 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.302 ± 0.023 | 2.291 ± 0.031 |
5.513 ± 0.03 | 3.043 ± 0.03 |
7.867 ± 0.051 | 4.689 ± 0.035 |
5.705 ± 0.06 | 7.483 ± 0.032 |
1.414 ± 0.021 | 2.209 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
