
Candidatus Purcelliella pentastirinorum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Enterobacteriaceae incertae sedis; ant, tsetse, mealybug, aphid, etc. endosymbionts; Candidatus Purcelliella
Average proteome isoelectric point is 8.6
Get precalculated fractions of proteins
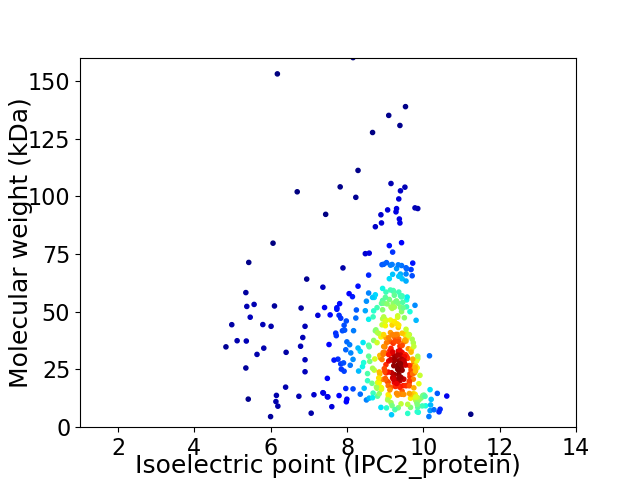
Virtual 2D-PAGE plot for 427 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
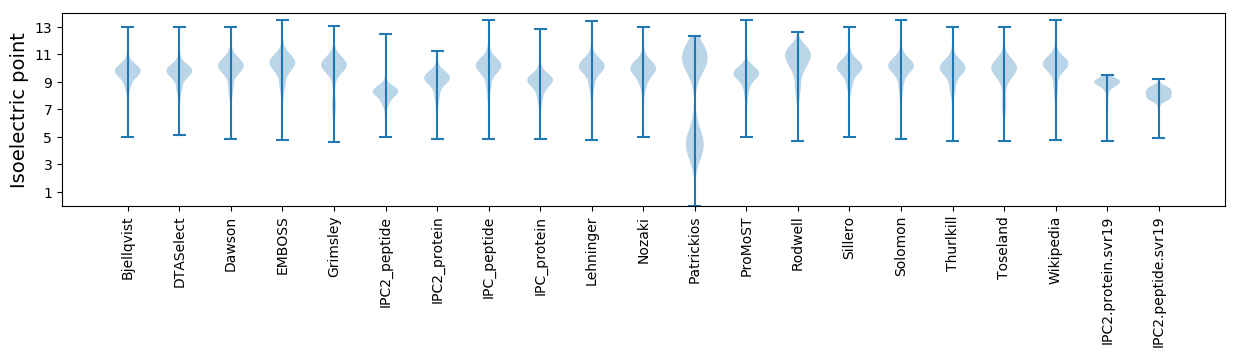
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346E091|A0A346E091_9ENTR Ribosomal RNA large subunit methyltransferase E OS=Candidatus Purcelliella pentastirinorum OX=472834 GN=C9I82_450 PE=4 SV=1
MM1 pKa = 7.38NKK3 pKa = 9.81IIFDD7 pKa = 3.91TDD9 pKa = 2.76IGIDD13 pKa = 3.55DD14 pKa = 4.68AFALLLAYY22 pKa = 7.58KK23 pKa = 8.02TQNILGITTVFGNTNINQVTNNAVLFSKK51 pKa = 10.61KK52 pKa = 9.82FDD54 pKa = 3.5IDD56 pKa = 3.58VPIYY60 pKa = 10.05KK61 pKa = 10.18GCSNSLVYY69 pKa = 10.92NNSFDD74 pKa = 4.53DD75 pKa = 4.48SINIHH80 pKa = 6.1GNNGLGDD87 pKa = 3.73IFNDD91 pKa = 3.3YY92 pKa = 10.67VNYY95 pKa = 9.89VAYY98 pKa = 10.12DD99 pKa = 3.47ALEE102 pKa = 4.49FIIDD106 pKa = 4.25SINDD110 pKa = 3.45NPHH113 pKa = 6.39EE114 pKa = 5.13IILVALGPLTNIATVIKK131 pKa = 9.96KK132 pKa = 10.44CPEE135 pKa = 3.95LVSKK139 pKa = 10.69IKK141 pKa = 10.44FLIIMGGAFGTDD153 pKa = 2.88GCFGNITCNSEE164 pKa = 3.87FNVWSDD170 pKa = 3.28PHH172 pKa = 6.22AAQLIFLSKK181 pKa = 10.67LPIVVVPLDD190 pKa = 3.66VTHH193 pKa = 7.1KK194 pKa = 10.64IVITEE199 pKa = 4.46DD200 pKa = 3.64DD201 pKa = 4.51LLSLNNNFLIDD212 pKa = 4.04LSRR215 pKa = 11.84CYY217 pKa = 10.42MSFYY221 pKa = 10.23RR222 pKa = 11.84NKK224 pKa = 10.37KK225 pKa = 9.6FFDD228 pKa = 3.46GMVLHH233 pKa = 7.05DD234 pKa = 5.79PIVISYY240 pKa = 10.22LIEE243 pKa = 4.96DD244 pKa = 3.22KK245 pKa = 10.88WFDD248 pKa = 3.59VKK250 pKa = 10.91DD251 pKa = 3.74VYY253 pKa = 11.67VEE255 pKa = 4.03VLTDD259 pKa = 3.24SLYY262 pKa = 10.72RR263 pKa = 11.84GSTLMYY269 pKa = 9.55DD270 pKa = 3.8KK271 pKa = 10.8KK272 pKa = 11.29YY273 pKa = 10.53DD274 pKa = 3.41FHH276 pKa = 8.32NILHH280 pKa = 6.73HH281 pKa = 6.74NDD283 pKa = 2.79SLLRR287 pKa = 11.84KK288 pKa = 9.3VCLNLDD294 pKa = 3.77VVNVKK299 pKa = 10.79SNLLSILKK307 pKa = 9.87II308 pKa = 3.86
MM1 pKa = 7.38NKK3 pKa = 9.81IIFDD7 pKa = 3.91TDD9 pKa = 2.76IGIDD13 pKa = 3.55DD14 pKa = 4.68AFALLLAYY22 pKa = 7.58KK23 pKa = 8.02TQNILGITTVFGNTNINQVTNNAVLFSKK51 pKa = 10.61KK52 pKa = 9.82FDD54 pKa = 3.5IDD56 pKa = 3.58VPIYY60 pKa = 10.05KK61 pKa = 10.18GCSNSLVYY69 pKa = 10.92NNSFDD74 pKa = 4.53DD75 pKa = 4.48SINIHH80 pKa = 6.1GNNGLGDD87 pKa = 3.73IFNDD91 pKa = 3.3YY92 pKa = 10.67VNYY95 pKa = 9.89VAYY98 pKa = 10.12DD99 pKa = 3.47ALEE102 pKa = 4.49FIIDD106 pKa = 4.25SINDD110 pKa = 3.45NPHH113 pKa = 6.39EE114 pKa = 5.13IILVALGPLTNIATVIKK131 pKa = 9.96KK132 pKa = 10.44CPEE135 pKa = 3.95LVSKK139 pKa = 10.69IKK141 pKa = 10.44FLIIMGGAFGTDD153 pKa = 2.88GCFGNITCNSEE164 pKa = 3.87FNVWSDD170 pKa = 3.28PHH172 pKa = 6.22AAQLIFLSKK181 pKa = 10.67LPIVVVPLDD190 pKa = 3.66VTHH193 pKa = 7.1KK194 pKa = 10.64IVITEE199 pKa = 4.46DD200 pKa = 3.64DD201 pKa = 4.51LLSLNNNFLIDD212 pKa = 4.04LSRR215 pKa = 11.84CYY217 pKa = 10.42MSFYY221 pKa = 10.23RR222 pKa = 11.84NKK224 pKa = 10.37KK225 pKa = 9.6FFDD228 pKa = 3.46GMVLHH233 pKa = 7.05DD234 pKa = 5.79PIVISYY240 pKa = 10.22LIEE243 pKa = 4.96DD244 pKa = 3.22KK245 pKa = 10.88WFDD248 pKa = 3.59VKK250 pKa = 10.91DD251 pKa = 3.74VYY253 pKa = 11.67VEE255 pKa = 4.03VLTDD259 pKa = 3.24SLYY262 pKa = 10.72RR263 pKa = 11.84GSTLMYY269 pKa = 9.55DD270 pKa = 3.8KK271 pKa = 10.8KK272 pKa = 11.29YY273 pKa = 10.53DD274 pKa = 3.41FHH276 pKa = 8.32NILHH280 pKa = 6.73HH281 pKa = 6.74NDD283 pKa = 2.79SLLRR287 pKa = 11.84KK288 pKa = 9.3VCLNLDD294 pKa = 3.77VVNVKK299 pKa = 10.79SNLLSILKK307 pKa = 9.87II308 pKa = 3.86
Molecular weight: 34.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346DZI5|A0A346DZI5_9ENTR 3'(2') 5'-bisphosphate nucleotidase CysQ OS=Candidatus Purcelliella pentastirinorum OX=472834 GN=cysQ PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.58RR3 pKa = 11.84TFQPSLIKK11 pKa = 10.35RR12 pKa = 11.84SRR14 pKa = 11.84LHH16 pKa = 6.02GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.97NGRR28 pKa = 11.84NILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84FKK37 pKa = 11.09GRR39 pKa = 11.84IRR41 pKa = 11.84LTVLWW46 pKa = 4.29
MM1 pKa = 7.45KK2 pKa = 9.58RR3 pKa = 11.84TFQPSLIKK11 pKa = 10.35RR12 pKa = 11.84SRR14 pKa = 11.84LHH16 pKa = 6.02GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.97NGRR28 pKa = 11.84NILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84FKK37 pKa = 11.09GRR39 pKa = 11.84IRR41 pKa = 11.84LTVLWW46 pKa = 4.29
Molecular weight: 5.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
139146 |
38 |
1428 |
325.9 |
37.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.525 ± 0.114 | 1.195 ± 0.041 |
3.991 ± 0.084 | 4.182 ± 0.119 |
5.341 ± 0.17 | 5.113 ± 0.116 |
1.634 ± 0.039 | 14.189 ± 0.195 |
11.729 ± 0.264 | 9.818 ± 0.131 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.163 ± 0.048 | 8.901 ± 0.17 |
2.62 ± 0.053 | 2.107 ± 0.058 |
3.32 ± 0.094 | 6.33 ± 0.101 |
4.13 ± 0.08 | 4.174 ± 0.123 |
0.908 ± 0.041 | 4.628 ± 0.091 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
