
Aureibaculum marinum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aureibaculum
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
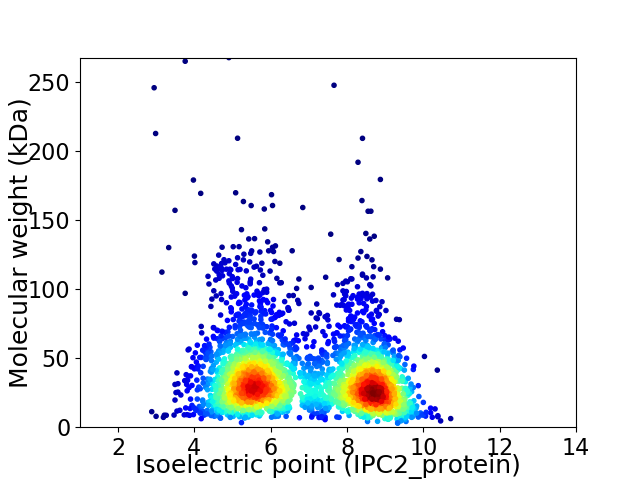
Virtual 2D-PAGE plot for 3096 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N4NWT2|A0A3N4NWT2_9FLAO Uncharacterized protein OS=Aureibaculum marinum OX=2487930 GN=EGM88_09820 PE=4 SV=1
MM1 pKa = 7.62IKK3 pKa = 10.14QFIKK7 pKa = 10.8LSTLCFVILLGFTSCDD23 pKa = 3.52SDD25 pKa = 4.09DD26 pKa = 4.37QDD28 pKa = 3.67PVMPKK33 pKa = 10.22GDD35 pKa = 3.72YY36 pKa = 9.26EE37 pKa = 3.93NGYY40 pKa = 9.98FVVNEE45 pKa = 4.41GNFGTPNGSVTFIDD59 pKa = 5.44DD60 pKa = 4.72NLLQEE65 pKa = 4.37NNEE68 pKa = 3.88IFQSVNSTTLGDD80 pKa = 3.58VVQYY84 pKa = 9.46ITFEE88 pKa = 4.33DD89 pKa = 4.41DD90 pKa = 3.26YY91 pKa = 11.95GFIVVNNSNKK101 pKa = 9.2IEE103 pKa = 4.15VVNRR107 pKa = 11.84YY108 pKa = 6.26TFEE111 pKa = 4.02SVATITEE118 pKa = 4.04NLQLPRR124 pKa = 11.84YY125 pKa = 9.44AVVEE129 pKa = 3.95NGKK132 pKa = 10.7LYY134 pKa = 9.18VTNSGTNSVEE144 pKa = 3.9VFDD147 pKa = 5.88ANTFAYY153 pKa = 9.6INSIDD158 pKa = 3.31IDD160 pKa = 3.81KK161 pKa = 9.81TVEE164 pKa = 4.2EE165 pKa = 4.31IVEE168 pKa = 4.36EE169 pKa = 4.0NDD171 pKa = 3.31LLYY174 pKa = 11.58VMNASFGFGNEE185 pKa = 3.17ITVINTQSDD194 pKa = 4.39TVVKK198 pKa = 9.62TITVGDD204 pKa = 3.95GLNSIEE210 pKa = 4.24VEE212 pKa = 4.12DD213 pKa = 5.56DD214 pKa = 2.78ILYY217 pKa = 10.3ALHH220 pKa = 6.6SAGITKK226 pKa = 10.17VNTATNEE233 pKa = 4.21VIGEE237 pKa = 4.23VPFEE241 pKa = 5.51DD242 pKa = 4.45GLLMASKK249 pKa = 11.11LEE251 pKa = 3.83VDD253 pKa = 3.68DD254 pKa = 4.61NYY256 pKa = 10.62IYY258 pKa = 10.61FLSGSKK264 pKa = 9.67IFKK267 pKa = 9.25YY268 pKa = 10.81NKK270 pKa = 10.04DD271 pKa = 3.41VTSLANTEE279 pKa = 4.39LVDD282 pKa = 3.92TQVEE286 pKa = 4.15DD287 pKa = 3.8ASWYY291 pKa = 9.39IGYY294 pKa = 9.7GFNVIDD300 pKa = 3.89NKK302 pKa = 10.85LFYY305 pKa = 10.32TDD307 pKa = 3.07VKK309 pKa = 10.85GFSEE313 pKa = 4.02NSEE316 pKa = 4.12VKK318 pKa = 10.75VYY320 pKa = 10.98DD321 pKa = 4.07LNGDD325 pKa = 4.05LLTTFNAGIGANGVYY340 pKa = 10.58EE341 pKa = 4.1NDD343 pKa = 3.3
MM1 pKa = 7.62IKK3 pKa = 10.14QFIKK7 pKa = 10.8LSTLCFVILLGFTSCDD23 pKa = 3.52SDD25 pKa = 4.09DD26 pKa = 4.37QDD28 pKa = 3.67PVMPKK33 pKa = 10.22GDD35 pKa = 3.72YY36 pKa = 9.26EE37 pKa = 3.93NGYY40 pKa = 9.98FVVNEE45 pKa = 4.41GNFGTPNGSVTFIDD59 pKa = 5.44DD60 pKa = 4.72NLLQEE65 pKa = 4.37NNEE68 pKa = 3.88IFQSVNSTTLGDD80 pKa = 3.58VVQYY84 pKa = 9.46ITFEE88 pKa = 4.33DD89 pKa = 4.41DD90 pKa = 3.26YY91 pKa = 11.95GFIVVNNSNKK101 pKa = 9.2IEE103 pKa = 4.15VVNRR107 pKa = 11.84YY108 pKa = 6.26TFEE111 pKa = 4.02SVATITEE118 pKa = 4.04NLQLPRR124 pKa = 11.84YY125 pKa = 9.44AVVEE129 pKa = 3.95NGKK132 pKa = 10.7LYY134 pKa = 9.18VTNSGTNSVEE144 pKa = 3.9VFDD147 pKa = 5.88ANTFAYY153 pKa = 9.6INSIDD158 pKa = 3.31IDD160 pKa = 3.81KK161 pKa = 9.81TVEE164 pKa = 4.2EE165 pKa = 4.31IVEE168 pKa = 4.36EE169 pKa = 4.0NDD171 pKa = 3.31LLYY174 pKa = 11.58VMNASFGFGNEE185 pKa = 3.17ITVINTQSDD194 pKa = 4.39TVVKK198 pKa = 9.62TITVGDD204 pKa = 3.95GLNSIEE210 pKa = 4.24VEE212 pKa = 4.12DD213 pKa = 5.56DD214 pKa = 2.78ILYY217 pKa = 10.3ALHH220 pKa = 6.6SAGITKK226 pKa = 10.17VNTATNEE233 pKa = 4.21VIGEE237 pKa = 4.23VPFEE241 pKa = 5.51DD242 pKa = 4.45GLLMASKK249 pKa = 11.11LEE251 pKa = 3.83VDD253 pKa = 3.68DD254 pKa = 4.61NYY256 pKa = 10.62IYY258 pKa = 10.61FLSGSKK264 pKa = 9.67IFKK267 pKa = 9.25YY268 pKa = 10.81NKK270 pKa = 10.04DD271 pKa = 3.41VTSLANTEE279 pKa = 4.39LVDD282 pKa = 3.92TQVEE286 pKa = 4.15DD287 pKa = 3.8ASWYY291 pKa = 9.39IGYY294 pKa = 9.7GFNVIDD300 pKa = 3.89NKK302 pKa = 10.85LFYY305 pKa = 10.32TDD307 pKa = 3.07VKK309 pKa = 10.85GFSEE313 pKa = 4.02NSEE316 pKa = 4.12VKK318 pKa = 10.75VYY320 pKa = 10.98DD321 pKa = 4.07LNGDD325 pKa = 4.05LLTTFNAGIGANGVYY340 pKa = 10.58EE341 pKa = 4.1NDD343 pKa = 3.3
Molecular weight: 38.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N4NTB4|A0A3N4NTB4_9FLAO Thioredoxin OS=Aureibaculum marinum OX=2487930 GN=EGM88_03280 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.05GFRR20 pKa = 11.84EE21 pKa = 4.13RR22 pKa = 11.84MSSVNGRR29 pKa = 11.84KK30 pKa = 9.11VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.76GRR40 pKa = 11.84KK41 pKa = 8.18RR42 pKa = 11.84LSVSSDD48 pKa = 3.33PSPKK52 pKa = 10.05KK53 pKa = 10.54
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.05GFRR20 pKa = 11.84EE21 pKa = 4.13RR22 pKa = 11.84MSSVNGRR29 pKa = 11.84KK30 pKa = 9.11VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.76GRR40 pKa = 11.84KK41 pKa = 8.18RR42 pKa = 11.84LSVSSDD48 pKa = 3.33PSPKK52 pKa = 10.05KK53 pKa = 10.54
Molecular weight: 6.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1089046 |
24 |
2458 |
351.8 |
39.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.867 ± 0.04 | 0.661 ± 0.012 |
5.578 ± 0.05 | 6.445 ± 0.039 |
5.122 ± 0.038 | 6.275 ± 0.053 |
1.7 ± 0.019 | 8.435 ± 0.043 |
8.57 ± 0.064 | 9.23 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.043 ± 0.019 | 6.746 ± 0.039 |
3.292 ± 0.023 | 3.176 ± 0.025 |
3.164 ± 0.024 | 6.506 ± 0.03 |
5.816 ± 0.041 | 6.062 ± 0.041 |
1.054 ± 0.017 | 4.26 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
