
Dolphin rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
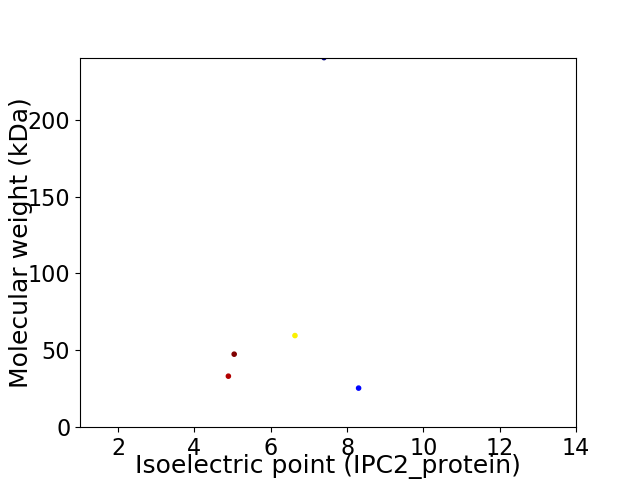
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A068EWX6|A0A068EWX6_9RHAB Nucleocapsid protein OS=Dolphin rhabdovirus OX=1511639 PE=4 SV=1
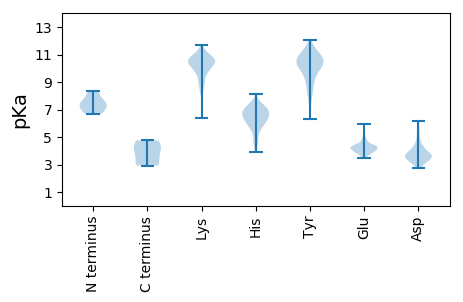
MM1 pKa = 7.14MMTRR5 pKa = 11.84RR6 pKa = 11.84PLKK9 pKa = 10.23IEE11 pKa = 3.74YY12 pKa = 11.04DD13 pKa = 3.48MDD15 pKa = 3.89KK16 pKa = 11.41LEE18 pKa = 4.79ASLKK22 pKa = 10.13EE23 pKa = 3.7ADD25 pKa = 4.39EE26 pKa = 4.5IEE28 pKa = 4.4GANEE32 pKa = 4.64GISEE36 pKa = 4.21RR37 pKa = 11.84MSDD40 pKa = 3.54LKK42 pKa = 11.17SLNEE46 pKa = 4.14SLDD49 pKa = 3.76DD50 pKa = 5.89LLAEE54 pKa = 5.43DD55 pKa = 5.14DD56 pKa = 3.62WAEE59 pKa = 3.85QGGIEE64 pKa = 4.29TDD66 pKa = 3.44AEE68 pKa = 4.33ADD70 pKa = 4.03DD71 pKa = 4.03EE72 pKa = 4.7HH73 pKa = 7.07EE74 pKa = 4.53SAVKK78 pKa = 10.73SSDD81 pKa = 2.88VDD83 pKa = 3.69MNPSDD88 pKa = 4.79DD89 pKa = 5.13AWDD92 pKa = 3.69SDD94 pKa = 4.5CFVPPDD100 pKa = 3.22IPGYY104 pKa = 6.29EE105 pKa = 3.83TRR107 pKa = 11.84GYY109 pKa = 10.46KK110 pKa = 10.17LSNSITPATRR120 pKa = 11.84LAILKK125 pKa = 8.22EE126 pKa = 3.85VSKK129 pKa = 10.63IIEE132 pKa = 4.37DD133 pKa = 3.61LRR135 pKa = 11.84GDD137 pKa = 3.6MVIAPKK143 pKa = 10.78GDD145 pKa = 3.26TYY147 pKa = 11.37DD148 pKa = 3.73YY149 pKa = 11.59YY150 pKa = 12.06LMFPDD155 pKa = 6.14GYY157 pKa = 11.05FNNKK161 pKa = 9.39DD162 pKa = 3.5VGRR165 pKa = 11.84KK166 pKa = 9.0HH167 pKa = 6.12YY168 pKa = 9.96PGDD171 pKa = 4.54RR172 pKa = 11.84IPPTDD177 pKa = 4.8DD178 pKa = 3.64PPGADD183 pKa = 3.64KK184 pKa = 10.82TPEE187 pKa = 3.91GLKK190 pKa = 8.85TRR192 pKa = 11.84KK193 pKa = 9.87NGANDD198 pKa = 3.47DD199 pKa = 3.78KK200 pKa = 11.34NRR202 pKa = 11.84RR203 pKa = 11.84SVKK206 pKa = 9.45EE207 pKa = 3.98ANATAQKK214 pKa = 10.38SNKK217 pKa = 8.81LASEE221 pKa = 3.58IWEE224 pKa = 4.22TGVVVIAKK232 pKa = 9.89DD233 pKa = 3.76GKK235 pKa = 10.59SKK237 pKa = 10.79LRR239 pKa = 11.84LKK241 pKa = 10.2PDD243 pKa = 2.93KK244 pKa = 10.78LYY246 pKa = 10.34WEE248 pKa = 4.0RR249 pKa = 11.84VEE251 pKa = 5.71WFKK254 pKa = 11.15QWDD257 pKa = 4.06EE258 pKa = 4.82KK259 pKa = 11.7NMEE262 pKa = 4.19TLPRR266 pKa = 11.84SKK268 pKa = 10.03IIQHH272 pKa = 7.0LIRR275 pKa = 11.84KK276 pKa = 6.84TPMRR280 pKa = 11.84LTYY283 pKa = 9.17RR284 pKa = 11.84RR285 pKa = 11.84KK286 pKa = 10.47YY287 pKa = 10.09ILEE290 pKa = 4.01
MM1 pKa = 7.14MMTRR5 pKa = 11.84RR6 pKa = 11.84PLKK9 pKa = 10.23IEE11 pKa = 3.74YY12 pKa = 11.04DD13 pKa = 3.48MDD15 pKa = 3.89KK16 pKa = 11.41LEE18 pKa = 4.79ASLKK22 pKa = 10.13EE23 pKa = 3.7ADD25 pKa = 4.39EE26 pKa = 4.5IEE28 pKa = 4.4GANEE32 pKa = 4.64GISEE36 pKa = 4.21RR37 pKa = 11.84MSDD40 pKa = 3.54LKK42 pKa = 11.17SLNEE46 pKa = 4.14SLDD49 pKa = 3.76DD50 pKa = 5.89LLAEE54 pKa = 5.43DD55 pKa = 5.14DD56 pKa = 3.62WAEE59 pKa = 3.85QGGIEE64 pKa = 4.29TDD66 pKa = 3.44AEE68 pKa = 4.33ADD70 pKa = 4.03DD71 pKa = 4.03EE72 pKa = 4.7HH73 pKa = 7.07EE74 pKa = 4.53SAVKK78 pKa = 10.73SSDD81 pKa = 2.88VDD83 pKa = 3.69MNPSDD88 pKa = 4.79DD89 pKa = 5.13AWDD92 pKa = 3.69SDD94 pKa = 4.5CFVPPDD100 pKa = 3.22IPGYY104 pKa = 6.29EE105 pKa = 3.83TRR107 pKa = 11.84GYY109 pKa = 10.46KK110 pKa = 10.17LSNSITPATRR120 pKa = 11.84LAILKK125 pKa = 8.22EE126 pKa = 3.85VSKK129 pKa = 10.63IIEE132 pKa = 4.37DD133 pKa = 3.61LRR135 pKa = 11.84GDD137 pKa = 3.6MVIAPKK143 pKa = 10.78GDD145 pKa = 3.26TYY147 pKa = 11.37DD148 pKa = 3.73YY149 pKa = 11.59YY150 pKa = 12.06LMFPDD155 pKa = 6.14GYY157 pKa = 11.05FNNKK161 pKa = 9.39DD162 pKa = 3.5VGRR165 pKa = 11.84KK166 pKa = 9.0HH167 pKa = 6.12YY168 pKa = 9.96PGDD171 pKa = 4.54RR172 pKa = 11.84IPPTDD177 pKa = 4.8DD178 pKa = 3.64PPGADD183 pKa = 3.64KK184 pKa = 10.82TPEE187 pKa = 3.91GLKK190 pKa = 8.85TRR192 pKa = 11.84KK193 pKa = 9.87NGANDD198 pKa = 3.47DD199 pKa = 3.78KK200 pKa = 11.34NRR202 pKa = 11.84RR203 pKa = 11.84SVKK206 pKa = 9.45EE207 pKa = 3.98ANATAQKK214 pKa = 10.38SNKK217 pKa = 8.81LASEE221 pKa = 3.58IWEE224 pKa = 4.22TGVVVIAKK232 pKa = 9.89DD233 pKa = 3.76GKK235 pKa = 10.59SKK237 pKa = 10.79LRR239 pKa = 11.84LKK241 pKa = 10.2PDD243 pKa = 2.93KK244 pKa = 10.78LYY246 pKa = 10.34WEE248 pKa = 4.0RR249 pKa = 11.84VEE251 pKa = 5.71WFKK254 pKa = 11.15QWDD257 pKa = 4.06EE258 pKa = 4.82KK259 pKa = 11.7NMEE262 pKa = 4.19TLPRR266 pKa = 11.84SKK268 pKa = 10.03IIQHH272 pKa = 7.0LIRR275 pKa = 11.84KK276 pKa = 6.84TPMRR280 pKa = 11.84LTYY283 pKa = 9.17RR284 pKa = 11.84RR285 pKa = 11.84KK286 pKa = 10.47YY287 pKa = 10.09ILEE290 pKa = 4.01
Molecular weight: 33.18 kDa
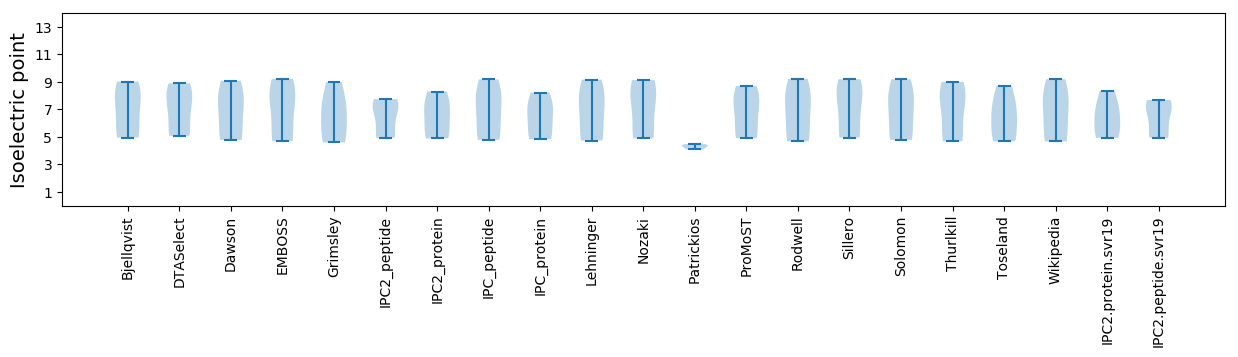
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A068EQH8|A0A068EQH8_9RHAB GDP polyribonucleotidyltransferase OS=Dolphin rhabdovirus OX=1511639 PE=4 SV=1
MM1 pKa = 7.39FKK3 pKa = 10.73SKK5 pKa = 10.66ALSLFRR11 pKa = 11.84KK12 pKa = 7.14TPKK15 pKa = 10.42DD16 pKa = 2.89KK17 pKa = 11.08TMNRR21 pKa = 11.84KK22 pKa = 9.52DD23 pKa = 3.3KK24 pKa = 11.22SPVKK28 pKa = 10.23RR29 pKa = 11.84VMKK32 pKa = 10.02YY33 pKa = 10.66NEE35 pKa = 4.07VADD38 pKa = 4.24SEE40 pKa = 5.31DD41 pKa = 3.61GFPLFKK47 pKa = 10.37PIPEE51 pKa = 4.05GTYY54 pKa = 9.98YY55 pKa = 10.8RR56 pKa = 11.84HH57 pKa = 6.04ITLKK61 pKa = 11.03VNFKK65 pKa = 10.78LSIVTKK71 pKa = 10.26EE72 pKa = 3.91PIEE75 pKa = 4.0NLMEE79 pKa = 4.62IYY81 pKa = 10.8LIGQHH86 pKa = 6.9ILDD89 pKa = 4.81NYY91 pKa = 10.0NGPARR96 pKa = 11.84SKK98 pKa = 10.81PMYY101 pKa = 9.21LALMIGGFNGLEE113 pKa = 4.23KK114 pKa = 10.51SHH116 pKa = 4.79QTSRR120 pKa = 11.84ITCYY124 pKa = 9.42EE125 pKa = 4.05RR126 pKa = 11.84EE127 pKa = 3.97FHH129 pKa = 7.0GPIDD133 pKa = 3.63FPYY136 pKa = 10.34YY137 pKa = 10.3RR138 pKa = 11.84RR139 pKa = 11.84DD140 pKa = 3.93PIDD143 pKa = 3.44WVIMPLSTAYY153 pKa = 8.54EE154 pKa = 4.37CSMKK158 pKa = 10.61GHH160 pKa = 6.36TEE162 pKa = 3.83VTVYY166 pKa = 11.25ASLTPTNMTGPSLAQFCEE184 pKa = 3.86GHH186 pKa = 5.84PRR188 pKa = 11.84MRR190 pKa = 11.84KK191 pKa = 8.98PPILDD196 pKa = 3.14TLTNFKK202 pKa = 10.31IDD204 pKa = 3.32TKK206 pKa = 10.92LKK208 pKa = 9.47NEE210 pKa = 3.92EE211 pKa = 3.58WHH213 pKa = 7.38LDD215 pKa = 3.42MSQCC219 pKa = 4.75
MM1 pKa = 7.39FKK3 pKa = 10.73SKK5 pKa = 10.66ALSLFRR11 pKa = 11.84KK12 pKa = 7.14TPKK15 pKa = 10.42DD16 pKa = 2.89KK17 pKa = 11.08TMNRR21 pKa = 11.84KK22 pKa = 9.52DD23 pKa = 3.3KK24 pKa = 11.22SPVKK28 pKa = 10.23RR29 pKa = 11.84VMKK32 pKa = 10.02YY33 pKa = 10.66NEE35 pKa = 4.07VADD38 pKa = 4.24SEE40 pKa = 5.31DD41 pKa = 3.61GFPLFKK47 pKa = 10.37PIPEE51 pKa = 4.05GTYY54 pKa = 9.98YY55 pKa = 10.8RR56 pKa = 11.84HH57 pKa = 6.04ITLKK61 pKa = 11.03VNFKK65 pKa = 10.78LSIVTKK71 pKa = 10.26EE72 pKa = 3.91PIEE75 pKa = 4.0NLMEE79 pKa = 4.62IYY81 pKa = 10.8LIGQHH86 pKa = 6.9ILDD89 pKa = 4.81NYY91 pKa = 10.0NGPARR96 pKa = 11.84SKK98 pKa = 10.81PMYY101 pKa = 9.21LALMIGGFNGLEE113 pKa = 4.23KK114 pKa = 10.51SHH116 pKa = 4.79QTSRR120 pKa = 11.84ITCYY124 pKa = 9.42EE125 pKa = 4.05RR126 pKa = 11.84EE127 pKa = 3.97FHH129 pKa = 7.0GPIDD133 pKa = 3.63FPYY136 pKa = 10.34YY137 pKa = 10.3RR138 pKa = 11.84RR139 pKa = 11.84DD140 pKa = 3.93PIDD143 pKa = 3.44WVIMPLSTAYY153 pKa = 8.54EE154 pKa = 4.37CSMKK158 pKa = 10.61GHH160 pKa = 6.36TEE162 pKa = 3.83VTVYY166 pKa = 11.25ASLTPTNMTGPSLAQFCEE184 pKa = 3.86GHH186 pKa = 5.84PRR188 pKa = 11.84MRR190 pKa = 11.84KK191 pKa = 8.98PPILDD196 pKa = 3.14TLTNFKK202 pKa = 10.31IDD204 pKa = 3.32TKK206 pKa = 10.92LKK208 pKa = 9.47NEE210 pKa = 3.92EE211 pKa = 3.58WHH213 pKa = 7.38LDD215 pKa = 3.42MSQCC219 pKa = 4.75
Molecular weight: 25.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3550 |
219 |
2086 |
710.0 |
81.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.62 ± 0.731 | 1.465 ± 0.275 |
5.859 ± 0.739 | 6.592 ± 0.648 |
4.0 ± 0.435 | 5.662 ± 0.402 |
2.535 ± 0.513 | 7.211 ± 0.404 |
6.873 ± 0.534 | 9.803 ± 1.114 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.268 ± 0.406 | 5.099 ± 0.517 |
4.704 ± 0.419 | 2.986 ± 0.299 |
5.296 ± 0.481 | 7.606 ± 0.316 |
5.775 ± 0.905 | 5.268 ± 0.426 |
1.746 ± 0.109 | 3.634 ± 0.173 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
