
Porcine stool-associated circular virus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
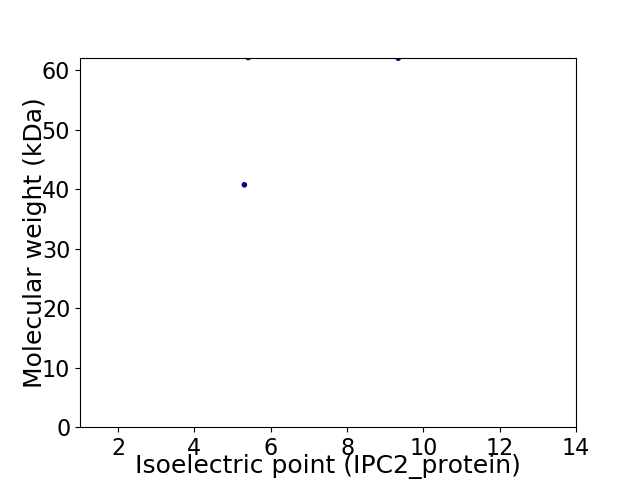
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8T0A7|W8T0A7_9CIRC Cap protein OS=Porcine stool-associated circular virus 5 OX=1475063 PE=4 SV=1
MM1 pKa = 7.83SSVRR5 pKa = 11.84LQNKK9 pKa = 9.18NLFLTYY15 pKa = 10.36NDD17 pKa = 3.9PEE19 pKa = 4.19EE20 pKa = 4.9TITGPAWLLNKK31 pKa = 10.0LAEE34 pKa = 4.23QLEE37 pKa = 4.83KK38 pKa = 10.02YY39 pKa = 8.97QPYY42 pKa = 10.49YY43 pKa = 10.31GVCGEE48 pKa = 4.56EE49 pKa = 4.09KK50 pKa = 10.87APTTGTKK57 pKa = 9.67HH58 pKa = 4.06YY59 pKa = 10.09HH60 pKa = 5.66LLIMCKK66 pKa = 10.25NPVISKK72 pKa = 9.11NGQVIEE78 pKa = 4.21IKK80 pKa = 10.74GIAPHH85 pKa = 6.66IEE87 pKa = 4.07KK88 pKa = 10.15IRR90 pKa = 11.84NNLRR94 pKa = 11.84GVIKK98 pKa = 10.24YY99 pKa = 9.02VKK101 pKa = 9.91KK102 pKa = 10.66DD103 pKa = 3.39GNIAEE108 pKa = 4.61LNKK111 pKa = 10.25NLCPVKK117 pKa = 10.35LDD119 pKa = 3.22KK120 pKa = 11.36LEE122 pKa = 4.45RR123 pKa = 11.84KK124 pKa = 9.25EE125 pKa = 4.62KK126 pKa = 10.59ADD128 pKa = 4.43LMLTGDD134 pKa = 4.24LEE136 pKa = 4.2KK137 pKa = 11.19AFIDD141 pKa = 3.97GTLGPIEE148 pKa = 4.36VIRR151 pKa = 11.84AEE153 pKa = 4.12KK154 pKa = 9.94LRR156 pKa = 11.84KK157 pKa = 9.13IFQVYY162 pKa = 9.66RR163 pKa = 11.84KK164 pKa = 9.41PEE166 pKa = 4.36PYY168 pKa = 9.81SKK170 pKa = 10.81KK171 pKa = 10.35LIMWYY176 pKa = 10.03RR177 pKa = 11.84GEE179 pKa = 4.09TGEE182 pKa = 4.3GKK184 pKa = 8.25TRR186 pKa = 11.84TAVEE190 pKa = 3.79LAEE193 pKa = 4.2EE194 pKa = 4.71FEE196 pKa = 4.13LSYY199 pKa = 10.49WITNEE204 pKa = 3.64SLRR207 pKa = 11.84WFDD210 pKa = 3.85GYY212 pKa = 11.34NGQEE216 pKa = 4.07LVIIDD221 pKa = 4.61DD222 pKa = 4.64FRR224 pKa = 11.84QKK226 pKa = 10.46MITDD230 pKa = 3.42WNFLLRR236 pKa = 11.84LLDD239 pKa = 4.3GYY241 pKa = 10.89GLSVPVKK248 pKa = 9.81GGYY251 pKa = 7.9VAWKK255 pKa = 8.61PKK257 pKa = 10.15IIIITTPATPQEE269 pKa = 4.04AFQWMNKK276 pKa = 10.08DD277 pKa = 3.78GEE279 pKa = 4.36LQNWDD284 pKa = 4.32HH285 pKa = 7.3LDD287 pKa = 3.28QLMRR291 pKa = 11.84RR292 pKa = 11.84LTHH295 pKa = 7.24DD296 pKa = 4.53DD297 pKa = 3.74EE298 pKa = 4.67EE299 pKa = 4.46QVYY302 pKa = 8.46QFPLWEE308 pKa = 4.12EE309 pKa = 4.29DD310 pKa = 3.27KK311 pKa = 11.09QKK313 pKa = 11.27LMNTVRR319 pKa = 11.84QFLGIEE325 pKa = 3.78NDD327 pKa = 3.17EE328 pKa = 4.33AMVEE332 pKa = 4.48EE333 pKa = 4.78EE334 pKa = 5.04LSMSPILPEE343 pKa = 3.62PTQIDD348 pKa = 3.83EE349 pKa = 4.25AA350 pKa = 5.31
MM1 pKa = 7.83SSVRR5 pKa = 11.84LQNKK9 pKa = 9.18NLFLTYY15 pKa = 10.36NDD17 pKa = 3.9PEE19 pKa = 4.19EE20 pKa = 4.9TITGPAWLLNKK31 pKa = 10.0LAEE34 pKa = 4.23QLEE37 pKa = 4.83KK38 pKa = 10.02YY39 pKa = 8.97QPYY42 pKa = 10.49YY43 pKa = 10.31GVCGEE48 pKa = 4.56EE49 pKa = 4.09KK50 pKa = 10.87APTTGTKK57 pKa = 9.67HH58 pKa = 4.06YY59 pKa = 10.09HH60 pKa = 5.66LLIMCKK66 pKa = 10.25NPVISKK72 pKa = 9.11NGQVIEE78 pKa = 4.21IKK80 pKa = 10.74GIAPHH85 pKa = 6.66IEE87 pKa = 4.07KK88 pKa = 10.15IRR90 pKa = 11.84NNLRR94 pKa = 11.84GVIKK98 pKa = 10.24YY99 pKa = 9.02VKK101 pKa = 9.91KK102 pKa = 10.66DD103 pKa = 3.39GNIAEE108 pKa = 4.61LNKK111 pKa = 10.25NLCPVKK117 pKa = 10.35LDD119 pKa = 3.22KK120 pKa = 11.36LEE122 pKa = 4.45RR123 pKa = 11.84KK124 pKa = 9.25EE125 pKa = 4.62KK126 pKa = 10.59ADD128 pKa = 4.43LMLTGDD134 pKa = 4.24LEE136 pKa = 4.2KK137 pKa = 11.19AFIDD141 pKa = 3.97GTLGPIEE148 pKa = 4.36VIRR151 pKa = 11.84AEE153 pKa = 4.12KK154 pKa = 9.94LRR156 pKa = 11.84KK157 pKa = 9.13IFQVYY162 pKa = 9.66RR163 pKa = 11.84KK164 pKa = 9.41PEE166 pKa = 4.36PYY168 pKa = 9.81SKK170 pKa = 10.81KK171 pKa = 10.35LIMWYY176 pKa = 10.03RR177 pKa = 11.84GEE179 pKa = 4.09TGEE182 pKa = 4.3GKK184 pKa = 8.25TRR186 pKa = 11.84TAVEE190 pKa = 3.79LAEE193 pKa = 4.2EE194 pKa = 4.71FEE196 pKa = 4.13LSYY199 pKa = 10.49WITNEE204 pKa = 3.64SLRR207 pKa = 11.84WFDD210 pKa = 3.85GYY212 pKa = 11.34NGQEE216 pKa = 4.07LVIIDD221 pKa = 4.61DD222 pKa = 4.64FRR224 pKa = 11.84QKK226 pKa = 10.46MITDD230 pKa = 3.42WNFLLRR236 pKa = 11.84LLDD239 pKa = 4.3GYY241 pKa = 10.89GLSVPVKK248 pKa = 9.81GGYY251 pKa = 7.9VAWKK255 pKa = 8.61PKK257 pKa = 10.15IIIITTPATPQEE269 pKa = 4.04AFQWMNKK276 pKa = 10.08DD277 pKa = 3.78GEE279 pKa = 4.36LQNWDD284 pKa = 4.32HH285 pKa = 7.3LDD287 pKa = 3.28QLMRR291 pKa = 11.84RR292 pKa = 11.84LTHH295 pKa = 7.24DD296 pKa = 4.53DD297 pKa = 3.74EE298 pKa = 4.67EE299 pKa = 4.46QVYY302 pKa = 8.46QFPLWEE308 pKa = 4.12EE309 pKa = 4.29DD310 pKa = 3.27KK311 pKa = 11.09QKK313 pKa = 11.27LMNTVRR319 pKa = 11.84QFLGIEE325 pKa = 3.78NDD327 pKa = 3.17EE328 pKa = 4.33AMVEE332 pKa = 4.48EE333 pKa = 4.78EE334 pKa = 5.04LSMSPILPEE343 pKa = 3.62PTQIDD348 pKa = 3.83EE349 pKa = 4.25AA350 pKa = 5.31
Molecular weight: 40.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8T0A7|W8T0A7_9CIRC Cap protein OS=Porcine stool-associated circular virus 5 OX=1475063 PE=4 SV=1
MM1 pKa = 7.66PKK3 pKa = 9.89RR4 pKa = 11.84YY5 pKa = 9.8ASRR8 pKa = 11.84KK9 pKa = 5.78RR10 pKa = 11.84TYY12 pKa = 9.69RR13 pKa = 11.84KK14 pKa = 9.43KK15 pKa = 10.03SRR17 pKa = 11.84KK18 pKa = 5.72ITRR21 pKa = 11.84FRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 8.68RR26 pKa = 11.84TGTRR30 pKa = 11.84AKK32 pKa = 9.35AKK34 pKa = 10.11SYY36 pKa = 10.48FRR38 pKa = 11.84LRR40 pKa = 11.84RR41 pKa = 11.84AAKK44 pKa = 9.98NYY46 pKa = 9.92SKK48 pKa = 10.73SRR50 pKa = 11.84KK51 pKa = 8.26IKK53 pKa = 9.43RR54 pKa = 11.84AKK56 pKa = 9.78RR57 pKa = 11.84QYY59 pKa = 10.92KK60 pKa = 7.73YY61 pKa = 9.22TAGKK65 pKa = 9.52GVKK68 pKa = 9.83NKK70 pKa = 9.79IKK72 pKa = 10.31RR73 pKa = 11.84LTMSEE78 pKa = 4.3FAQLPFYY85 pKa = 10.84DD86 pKa = 3.63IARR89 pKa = 11.84LSSMVMSGGNPNMYY103 pKa = 10.26PNIPDD108 pKa = 4.36NIRR111 pKa = 11.84SAYY114 pKa = 9.27KK115 pKa = 10.64GQFGPYY121 pKa = 6.78WSRR124 pKa = 11.84YY125 pKa = 7.41GAILPEE131 pKa = 4.44EE132 pKa = 4.42YY133 pKa = 10.51FNASSPYY140 pKa = 10.63IMGNANDD147 pKa = 4.04SYY149 pKa = 11.43TPYY152 pKa = 10.69LYY154 pKa = 10.76AFNPILSPVTCGSKK168 pKa = 8.47MQLYY172 pKa = 9.64AALYY176 pKa = 10.39KK177 pKa = 10.13KK178 pKa = 10.69FKK180 pKa = 10.4YY181 pKa = 10.55VGIKK185 pKa = 10.23VSWHH189 pKa = 6.36PLAKK193 pKa = 9.04PTTTVVSTSMTVNPGMHH210 pKa = 7.24DD211 pKa = 3.4EE212 pKa = 4.71TYY214 pKa = 10.02QAYY217 pKa = 10.72SIDD220 pKa = 3.51AGKK223 pKa = 10.63KK224 pKa = 8.66VDD226 pKa = 3.35SFMNSGTTQGLYY238 pKa = 10.41LKK240 pKa = 9.67HH241 pKa = 7.04NSMEE245 pKa = 4.29RR246 pKa = 11.84NVRR249 pKa = 11.84AKK251 pKa = 10.81DD252 pKa = 4.31FIPMSTQTLYY262 pKa = 9.1MHH264 pKa = 6.96VYY266 pKa = 9.62FGKK269 pKa = 10.48QIEE272 pKa = 4.7SSQNWHH278 pKa = 6.65LPFINKK284 pKa = 9.09VSEE287 pKa = 4.2TGGVTGYY294 pKa = 11.17AVAHH298 pKa = 6.95GINDD302 pKa = 3.57NQRR305 pKa = 11.84GRR307 pKa = 11.84LKK309 pKa = 10.56AAPFYY314 pKa = 10.4DD315 pKa = 3.81IKK317 pKa = 10.61TMMPCIEE324 pKa = 4.87RR325 pKa = 11.84DD326 pKa = 3.6SKK328 pKa = 10.51LHH330 pKa = 6.04KK331 pKa = 10.49VYY333 pKa = 11.14DD334 pKa = 3.55MSKK337 pKa = 8.64PFSFFVRR344 pKa = 11.84PMVSSNEE351 pKa = 3.85VSKK354 pKa = 10.86EE355 pKa = 3.46AGINVTQEE363 pKa = 3.65AGEE366 pKa = 4.83LLTQPTNEE374 pKa = 4.02IEE376 pKa = 4.05QLQKK380 pKa = 10.97GLKK383 pKa = 8.77HH384 pKa = 6.32LGYY387 pKa = 10.55KK388 pKa = 10.2EE389 pKa = 4.13FTHH392 pKa = 6.75SLTSNKK398 pKa = 9.84LDD400 pKa = 3.51IPQDD404 pKa = 3.43AYY406 pKa = 11.18SAQHH410 pKa = 6.1AHH412 pKa = 7.07DD413 pKa = 4.72PQDD416 pKa = 3.1KK417 pKa = 10.24FYY419 pKa = 10.78HH420 pKa = 5.92RR421 pKa = 11.84WSALSDD427 pKa = 3.46DD428 pKa = 5.38DD429 pKa = 5.66NFFNPVMFWYY439 pKa = 10.57CFTTSDD445 pKa = 4.33RR446 pKa = 11.84YY447 pKa = 9.63VPTYY451 pKa = 10.66AEE453 pKa = 3.96YY454 pKa = 9.66MKK456 pKa = 10.84INDD459 pKa = 3.48PHH461 pKa = 7.7QLFNTNRR468 pKa = 11.84FFTTTGEE475 pKa = 4.4SNNAKK480 pKa = 10.16YY481 pKa = 10.04EE482 pKa = 3.86LSARR486 pKa = 11.84IDD488 pKa = 3.35TSAPFLNGYY497 pKa = 10.22GYY499 pKa = 11.11FDD501 pKa = 3.18VTFYY505 pKa = 10.86TKK507 pKa = 9.77WKK509 pKa = 9.49EE510 pKa = 3.75EE511 pKa = 3.93RR512 pKa = 11.84PLVSKK517 pKa = 9.58TSYY520 pKa = 10.59VGIVPSEE527 pKa = 4.08TGVEE531 pKa = 4.47GVSSIIGNN539 pKa = 3.69
MM1 pKa = 7.66PKK3 pKa = 9.89RR4 pKa = 11.84YY5 pKa = 9.8ASRR8 pKa = 11.84KK9 pKa = 5.78RR10 pKa = 11.84TYY12 pKa = 9.69RR13 pKa = 11.84KK14 pKa = 9.43KK15 pKa = 10.03SRR17 pKa = 11.84KK18 pKa = 5.72ITRR21 pKa = 11.84FRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 8.68RR26 pKa = 11.84TGTRR30 pKa = 11.84AKK32 pKa = 9.35AKK34 pKa = 10.11SYY36 pKa = 10.48FRR38 pKa = 11.84LRR40 pKa = 11.84RR41 pKa = 11.84AAKK44 pKa = 9.98NYY46 pKa = 9.92SKK48 pKa = 10.73SRR50 pKa = 11.84KK51 pKa = 8.26IKK53 pKa = 9.43RR54 pKa = 11.84AKK56 pKa = 9.78RR57 pKa = 11.84QYY59 pKa = 10.92KK60 pKa = 7.73YY61 pKa = 9.22TAGKK65 pKa = 9.52GVKK68 pKa = 9.83NKK70 pKa = 9.79IKK72 pKa = 10.31RR73 pKa = 11.84LTMSEE78 pKa = 4.3FAQLPFYY85 pKa = 10.84DD86 pKa = 3.63IARR89 pKa = 11.84LSSMVMSGGNPNMYY103 pKa = 10.26PNIPDD108 pKa = 4.36NIRR111 pKa = 11.84SAYY114 pKa = 9.27KK115 pKa = 10.64GQFGPYY121 pKa = 6.78WSRR124 pKa = 11.84YY125 pKa = 7.41GAILPEE131 pKa = 4.44EE132 pKa = 4.42YY133 pKa = 10.51FNASSPYY140 pKa = 10.63IMGNANDD147 pKa = 4.04SYY149 pKa = 11.43TPYY152 pKa = 10.69LYY154 pKa = 10.76AFNPILSPVTCGSKK168 pKa = 8.47MQLYY172 pKa = 9.64AALYY176 pKa = 10.39KK177 pKa = 10.13KK178 pKa = 10.69FKK180 pKa = 10.4YY181 pKa = 10.55VGIKK185 pKa = 10.23VSWHH189 pKa = 6.36PLAKK193 pKa = 9.04PTTTVVSTSMTVNPGMHH210 pKa = 7.24DD211 pKa = 3.4EE212 pKa = 4.71TYY214 pKa = 10.02QAYY217 pKa = 10.72SIDD220 pKa = 3.51AGKK223 pKa = 10.63KK224 pKa = 8.66VDD226 pKa = 3.35SFMNSGTTQGLYY238 pKa = 10.41LKK240 pKa = 9.67HH241 pKa = 7.04NSMEE245 pKa = 4.29RR246 pKa = 11.84NVRR249 pKa = 11.84AKK251 pKa = 10.81DD252 pKa = 4.31FIPMSTQTLYY262 pKa = 9.1MHH264 pKa = 6.96VYY266 pKa = 9.62FGKK269 pKa = 10.48QIEE272 pKa = 4.7SSQNWHH278 pKa = 6.65LPFINKK284 pKa = 9.09VSEE287 pKa = 4.2TGGVTGYY294 pKa = 11.17AVAHH298 pKa = 6.95GINDD302 pKa = 3.57NQRR305 pKa = 11.84GRR307 pKa = 11.84LKK309 pKa = 10.56AAPFYY314 pKa = 10.4DD315 pKa = 3.81IKK317 pKa = 10.61TMMPCIEE324 pKa = 4.87RR325 pKa = 11.84DD326 pKa = 3.6SKK328 pKa = 10.51LHH330 pKa = 6.04KK331 pKa = 10.49VYY333 pKa = 11.14DD334 pKa = 3.55MSKK337 pKa = 8.64PFSFFVRR344 pKa = 11.84PMVSSNEE351 pKa = 3.85VSKK354 pKa = 10.86EE355 pKa = 3.46AGINVTQEE363 pKa = 3.65AGEE366 pKa = 4.83LLTQPTNEE374 pKa = 4.02IEE376 pKa = 4.05QLQKK380 pKa = 10.97GLKK383 pKa = 8.77HH384 pKa = 6.32LGYY387 pKa = 10.55KK388 pKa = 10.2EE389 pKa = 4.13FTHH392 pKa = 6.75SLTSNKK398 pKa = 9.84LDD400 pKa = 3.51IPQDD404 pKa = 3.43AYY406 pKa = 11.18SAQHH410 pKa = 6.1AHH412 pKa = 7.07DD413 pKa = 4.72PQDD416 pKa = 3.1KK417 pKa = 10.24FYY419 pKa = 10.78HH420 pKa = 5.92RR421 pKa = 11.84WSALSDD427 pKa = 3.46DD428 pKa = 5.38DD429 pKa = 5.66NFFNPVMFWYY439 pKa = 10.57CFTTSDD445 pKa = 4.33RR446 pKa = 11.84YY447 pKa = 9.63VPTYY451 pKa = 10.66AEE453 pKa = 3.96YY454 pKa = 9.66MKK456 pKa = 10.84INDD459 pKa = 3.48PHH461 pKa = 7.7QLFNTNRR468 pKa = 11.84FFTTTGEE475 pKa = 4.4SNNAKK480 pKa = 10.16YY481 pKa = 10.04EE482 pKa = 3.86LSARR486 pKa = 11.84IDD488 pKa = 3.35TSAPFLNGYY497 pKa = 10.22GYY499 pKa = 11.11FDD501 pKa = 3.18VTFYY505 pKa = 10.86TKK507 pKa = 9.77WKK509 pKa = 9.49EE510 pKa = 3.75EE511 pKa = 3.93RR512 pKa = 11.84PLVSKK517 pKa = 9.58TSYY520 pKa = 10.59VGIVPSEE527 pKa = 4.08TGVEE531 pKa = 4.47GVSSIIGNN539 pKa = 3.69
Molecular weight: 61.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
889 |
350 |
539 |
444.5 |
51.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.512 ± 0.728 | 0.675 ± 0.108 |
4.612 ± 0.485 | 6.524 ± 2.233 |
4.274 ± 0.841 | 6.074 ± 0.126 |
2.025 ± 0.354 | 5.737 ± 1.004 |
8.661 ± 0.116 | 7.424 ± 2.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.262 ± 0.24 | 5.512 ± 0.219 |
5.287 ± 0.085 | 3.825 ± 0.443 |
5.174 ± 0.528 | 6.299 ± 2.213 |
6.412 ± 0.584 | 4.949 ± 0.055 |
1.687 ± 0.525 | 6.074 ± 1.231 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
