
Beihai sobemo-like virus 25
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
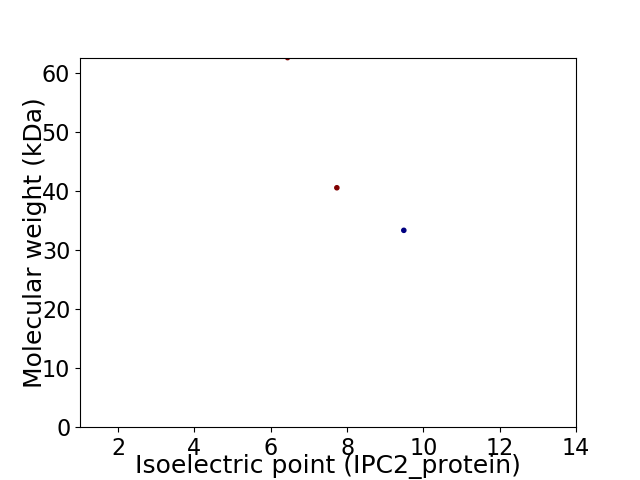
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE85|A0A1L3KE85_9VIRU Capsid protein OS=Beihai sobemo-like virus 25 OX=1922697 PE=3 SV=1
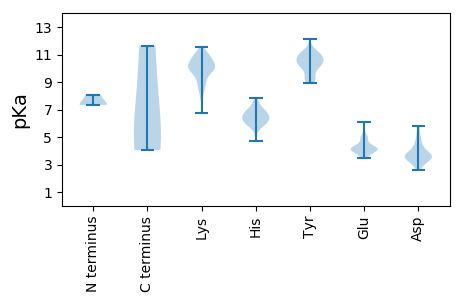
MM1 pKa = 8.07DD2 pKa = 4.1SMGPRR7 pKa = 11.84WRR9 pKa = 11.84SKK11 pKa = 7.68QQYY14 pKa = 10.29VDD16 pKa = 4.48FDD18 pKa = 4.52DD19 pKa = 5.36DD20 pKa = 4.22RR21 pKa = 11.84YY22 pKa = 10.75HH23 pKa = 6.1RR24 pKa = 11.84HH25 pKa = 5.54SYY27 pKa = 10.27EE28 pKa = 3.77NASIAGSVTRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84SSKK44 pKa = 10.55KK45 pKa = 8.74EE46 pKa = 3.64RR47 pKa = 11.84QRR49 pKa = 11.84PATKK53 pKa = 10.08DD54 pKa = 3.14RR55 pKa = 11.84PSEE58 pKa = 3.8VHH60 pKa = 5.84YY61 pKa = 10.78KK62 pKa = 10.15IVGKK66 pKa = 10.04GKK68 pKa = 9.17PVHH71 pKa = 6.34GPSAPKK77 pKa = 8.85MQPEE81 pKa = 4.24ALQVIEE87 pKa = 4.19DD88 pKa = 3.76HH89 pKa = 6.67KK90 pKa = 11.45EE91 pKa = 3.75EE92 pKa = 4.02IVALGFEE99 pKa = 4.62EE100 pKa = 6.09GVFAYY105 pKa = 9.97PDD107 pKa = 3.53MSPDD111 pKa = 3.58TEE113 pKa = 4.14RR114 pKa = 11.84KK115 pKa = 9.92SLEE118 pKa = 3.45AHH120 pKa = 6.26LNLFGEE126 pKa = 4.6RR127 pKa = 11.84TRR129 pKa = 11.84TITHH133 pKa = 7.32PPTDD137 pKa = 4.75DD138 pKa = 2.87EE139 pKa = 4.42HH140 pKa = 6.72QRR142 pKa = 11.84CSRR145 pKa = 11.84LVAEE149 pKa = 5.1MMQSATFVPKK159 pKa = 9.87SDD161 pKa = 3.36YY162 pKa = 11.06RR163 pKa = 11.84EE164 pKa = 3.82VSGVLDD170 pKa = 4.93IINSSIIDD178 pKa = 3.6PKK180 pKa = 10.44KK181 pKa = 9.99ASGYY185 pKa = 9.15PYY187 pKa = 10.26CLEE190 pKa = 4.21GKK192 pKa = 6.76PTNGQVLEE200 pKa = 4.2QYY202 pKa = 9.98GVRR205 pKa = 11.84GFAQHH210 pKa = 6.78VLNTWEE216 pKa = 4.77DD217 pKa = 3.68LPFQVKK223 pKa = 10.13NFLKK227 pKa = 10.9GEE229 pKa = 4.12PTKK232 pKa = 9.97KK233 pKa = 8.99TKK235 pKa = 10.23LDD237 pKa = 3.32KK238 pKa = 11.21GMPRR242 pKa = 11.84CIEE245 pKa = 4.11GFPLHH250 pKa = 5.84VTVKK254 pKa = 9.2HH255 pKa = 5.93ASVFSQLAMTLVKK268 pKa = 9.55QWKK271 pKa = 8.96HH272 pKa = 5.78IPVKK276 pKa = 10.68YY277 pKa = 10.31AFSPANPGHH286 pKa = 6.41IEE288 pKa = 3.92HH289 pKa = 7.0LKK291 pKa = 9.61EE292 pKa = 4.06CLPGKK297 pKa = 9.85VWEE300 pKa = 4.24SDD302 pKa = 3.18KK303 pKa = 11.55TNWDD307 pKa = 3.36YY308 pKa = 12.12LMYY311 pKa = 10.64LWIANVVRR319 pKa = 11.84DD320 pKa = 4.24SIKK323 pKa = 10.83KK324 pKa = 10.05LVIKK328 pKa = 9.53PAEE331 pKa = 4.09WTEE334 pKa = 4.01EE335 pKa = 3.96QYY337 pKa = 11.24SIYY340 pKa = 10.68LSDD343 pKa = 3.76IDD345 pKa = 4.51GCFKK349 pKa = 10.78QVFEE353 pKa = 4.21EE354 pKa = 4.08ASYY357 pKa = 8.92RR358 pKa = 11.84TSDD361 pKa = 3.11GHH363 pKa = 7.85IYY365 pKa = 10.26QSNEE369 pKa = 3.45PGIMKK374 pKa = 10.14SGWFMTIGANSIAQVAVHH392 pKa = 6.23VMTCIRR398 pKa = 11.84LGMSDD403 pKa = 5.61DD404 pKa = 5.81DD405 pKa = 4.1ILSTPIVAGGDD416 pKa = 3.73DD417 pKa = 4.03VNQAPVPAGKK427 pKa = 9.68AAYY430 pKa = 10.15LEE432 pKa = 4.34EE433 pKa = 4.13AQKK436 pKa = 11.0LGVSMEE442 pKa = 3.54IHH444 pKa = 6.11EE445 pKa = 5.11RR446 pKa = 11.84GDD448 pKa = 3.66LYY450 pKa = 10.89EE451 pKa = 4.67SEE453 pKa = 4.74YY454 pKa = 10.84FSSDD458 pKa = 2.6LRR460 pKa = 11.84LGVEE464 pKa = 4.35GPEE467 pKa = 4.05FFPKK471 pKa = 9.81RR472 pKa = 11.84WTKK475 pKa = 10.53HH476 pKa = 4.69IEE478 pKa = 3.67HH479 pKa = 7.14LKK481 pKa = 7.98TVKK484 pKa = 10.45RR485 pKa = 11.84EE486 pKa = 3.98NLADD490 pKa = 3.64ALVSHH495 pKa = 6.5MEE497 pKa = 4.13NYY499 pKa = 9.33RR500 pKa = 11.84HH501 pKa = 6.52HH502 pKa = 6.92VDD504 pKa = 4.17KK505 pKa = 11.24FNLLVKK511 pKa = 10.43LYY513 pKa = 10.89LSLEE517 pKa = 4.21EE518 pKa = 5.22KK519 pKa = 10.93YY520 pKa = 10.43PADD523 pKa = 3.77FPKK526 pKa = 10.76SKK528 pKa = 10.38LVSRR532 pKa = 11.84SLLRR536 pKa = 11.84ARR538 pKa = 11.84QYY540 pKa = 11.25GYY542 pKa = 10.94EE543 pKa = 4.03HH544 pKa = 7.46ALLCC548 pKa = 5.34
MM1 pKa = 8.07DD2 pKa = 4.1SMGPRR7 pKa = 11.84WRR9 pKa = 11.84SKK11 pKa = 7.68QQYY14 pKa = 10.29VDD16 pKa = 4.48FDD18 pKa = 4.52DD19 pKa = 5.36DD20 pKa = 4.22RR21 pKa = 11.84YY22 pKa = 10.75HH23 pKa = 6.1RR24 pKa = 11.84HH25 pKa = 5.54SYY27 pKa = 10.27EE28 pKa = 3.77NASIAGSVTRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84SSKK44 pKa = 10.55KK45 pKa = 8.74EE46 pKa = 3.64RR47 pKa = 11.84QRR49 pKa = 11.84PATKK53 pKa = 10.08DD54 pKa = 3.14RR55 pKa = 11.84PSEE58 pKa = 3.8VHH60 pKa = 5.84YY61 pKa = 10.78KK62 pKa = 10.15IVGKK66 pKa = 10.04GKK68 pKa = 9.17PVHH71 pKa = 6.34GPSAPKK77 pKa = 8.85MQPEE81 pKa = 4.24ALQVIEE87 pKa = 4.19DD88 pKa = 3.76HH89 pKa = 6.67KK90 pKa = 11.45EE91 pKa = 3.75EE92 pKa = 4.02IVALGFEE99 pKa = 4.62EE100 pKa = 6.09GVFAYY105 pKa = 9.97PDD107 pKa = 3.53MSPDD111 pKa = 3.58TEE113 pKa = 4.14RR114 pKa = 11.84KK115 pKa = 9.92SLEE118 pKa = 3.45AHH120 pKa = 6.26LNLFGEE126 pKa = 4.6RR127 pKa = 11.84TRR129 pKa = 11.84TITHH133 pKa = 7.32PPTDD137 pKa = 4.75DD138 pKa = 2.87EE139 pKa = 4.42HH140 pKa = 6.72QRR142 pKa = 11.84CSRR145 pKa = 11.84LVAEE149 pKa = 5.1MMQSATFVPKK159 pKa = 9.87SDD161 pKa = 3.36YY162 pKa = 11.06RR163 pKa = 11.84EE164 pKa = 3.82VSGVLDD170 pKa = 4.93IINSSIIDD178 pKa = 3.6PKK180 pKa = 10.44KK181 pKa = 9.99ASGYY185 pKa = 9.15PYY187 pKa = 10.26CLEE190 pKa = 4.21GKK192 pKa = 6.76PTNGQVLEE200 pKa = 4.2QYY202 pKa = 9.98GVRR205 pKa = 11.84GFAQHH210 pKa = 6.78VLNTWEE216 pKa = 4.77DD217 pKa = 3.68LPFQVKK223 pKa = 10.13NFLKK227 pKa = 10.9GEE229 pKa = 4.12PTKK232 pKa = 9.97KK233 pKa = 8.99TKK235 pKa = 10.23LDD237 pKa = 3.32KK238 pKa = 11.21GMPRR242 pKa = 11.84CIEE245 pKa = 4.11GFPLHH250 pKa = 5.84VTVKK254 pKa = 9.2HH255 pKa = 5.93ASVFSQLAMTLVKK268 pKa = 9.55QWKK271 pKa = 8.96HH272 pKa = 5.78IPVKK276 pKa = 10.68YY277 pKa = 10.31AFSPANPGHH286 pKa = 6.41IEE288 pKa = 3.92HH289 pKa = 7.0LKK291 pKa = 9.61EE292 pKa = 4.06CLPGKK297 pKa = 9.85VWEE300 pKa = 4.24SDD302 pKa = 3.18KK303 pKa = 11.55TNWDD307 pKa = 3.36YY308 pKa = 12.12LMYY311 pKa = 10.64LWIANVVRR319 pKa = 11.84DD320 pKa = 4.24SIKK323 pKa = 10.83KK324 pKa = 10.05LVIKK328 pKa = 9.53PAEE331 pKa = 4.09WTEE334 pKa = 4.01EE335 pKa = 3.96QYY337 pKa = 11.24SIYY340 pKa = 10.68LSDD343 pKa = 3.76IDD345 pKa = 4.51GCFKK349 pKa = 10.78QVFEE353 pKa = 4.21EE354 pKa = 4.08ASYY357 pKa = 8.92RR358 pKa = 11.84TSDD361 pKa = 3.11GHH363 pKa = 7.85IYY365 pKa = 10.26QSNEE369 pKa = 3.45PGIMKK374 pKa = 10.14SGWFMTIGANSIAQVAVHH392 pKa = 6.23VMTCIRR398 pKa = 11.84LGMSDD403 pKa = 5.61DD404 pKa = 5.81DD405 pKa = 4.1ILSTPIVAGGDD416 pKa = 3.73DD417 pKa = 4.03VNQAPVPAGKK427 pKa = 9.68AAYY430 pKa = 10.15LEE432 pKa = 4.34EE433 pKa = 4.13AQKK436 pKa = 11.0LGVSMEE442 pKa = 3.54IHH444 pKa = 6.11EE445 pKa = 5.11RR446 pKa = 11.84GDD448 pKa = 3.66LYY450 pKa = 10.89EE451 pKa = 4.67SEE453 pKa = 4.74YY454 pKa = 10.84FSSDD458 pKa = 2.6LRR460 pKa = 11.84LGVEE464 pKa = 4.35GPEE467 pKa = 4.05FFPKK471 pKa = 9.81RR472 pKa = 11.84WTKK475 pKa = 10.53HH476 pKa = 4.69IEE478 pKa = 3.67HH479 pKa = 7.14LKK481 pKa = 7.98TVKK484 pKa = 10.45RR485 pKa = 11.84EE486 pKa = 3.98NLADD490 pKa = 3.64ALVSHH495 pKa = 6.5MEE497 pKa = 4.13NYY499 pKa = 9.33RR500 pKa = 11.84HH501 pKa = 6.52HH502 pKa = 6.92VDD504 pKa = 4.17KK505 pKa = 11.24FNLLVKK511 pKa = 10.43LYY513 pKa = 10.89LSLEE517 pKa = 4.21EE518 pKa = 5.22KK519 pKa = 10.93YY520 pKa = 10.43PADD523 pKa = 3.77FPKK526 pKa = 10.76SKK528 pKa = 10.38LVSRR532 pKa = 11.84SLLRR536 pKa = 11.84ARR538 pKa = 11.84QYY540 pKa = 11.25GYY542 pKa = 10.94EE543 pKa = 4.03HH544 pKa = 7.46ALLCC548 pKa = 5.34
Molecular weight: 62.65 kDa
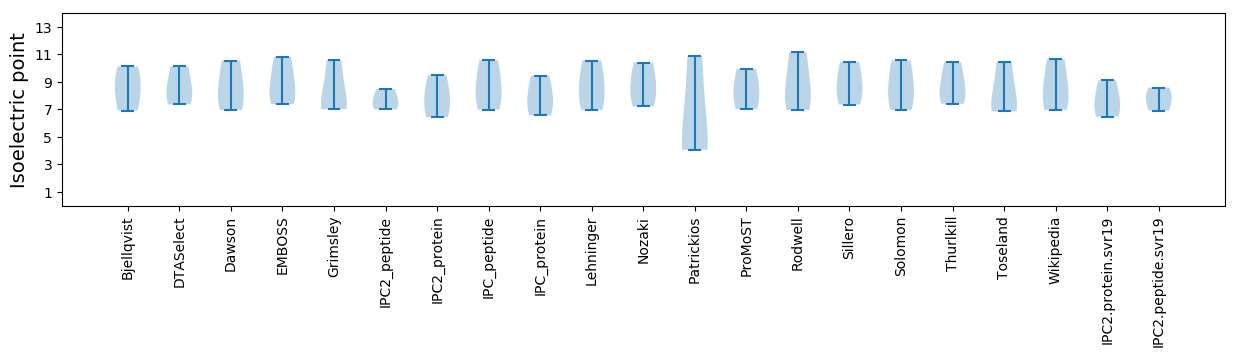
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE90|A0A1L3KE90_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 25 OX=1922697 PE=4 SV=1
MM1 pKa = 7.34NNSLTTQYY9 pKa = 11.32LGIPGEE15 pKa = 5.02DD16 pKa = 3.5PTSPPWASGNYY27 pKa = 9.33VGPFWSNGKK36 pKa = 8.68LQEE39 pKa = 4.24SVEE42 pKa = 4.39FGDD45 pKa = 5.04APPLHH50 pKa = 6.74EE51 pKa = 5.65LDD53 pKa = 3.66ALARR57 pKa = 11.84LHH59 pKa = 6.09DD60 pKa = 3.76TAYY63 pKa = 10.78ARR65 pKa = 11.84YY66 pKa = 9.22KK67 pKa = 10.92DD68 pKa = 3.75SAHH71 pKa = 6.8RR72 pKa = 11.84AAADD76 pKa = 3.37EE77 pKa = 4.36LFAEE81 pKa = 4.78EE82 pKa = 5.04AEE84 pKa = 4.32KK85 pKa = 10.89LKK87 pKa = 10.89KK88 pKa = 10.35KK89 pKa = 10.68YY90 pKa = 10.15GPNWAGNPQVAAKK103 pKa = 9.91LVRR106 pKa = 11.84YY107 pKa = 9.28GNHH110 pKa = 6.51TIRR113 pKa = 11.84AASRR117 pKa = 11.84VGGNVATGFKK127 pKa = 10.61FGGPLGAVGGLLYY140 pKa = 10.92SGLQNIKK147 pKa = 9.87QSHH150 pKa = 6.48DD151 pKa = 3.62MVTGNYY157 pKa = 9.02LRR159 pKa = 11.84KK160 pKa = 9.37EE161 pKa = 4.12RR162 pKa = 11.84KK163 pKa = 9.52DD164 pKa = 3.55LLSLFSKK171 pKa = 10.99DD172 pKa = 3.07PMKK175 pKa = 10.28TIKK178 pKa = 9.94VQPIGGSVVEE188 pKa = 4.93PGSSDD193 pKa = 3.42SKK195 pKa = 10.37PKK197 pKa = 10.64KK198 pKa = 8.52KK199 pKa = 8.95TLVEE203 pKa = 3.94KK204 pKa = 10.78LKK206 pKa = 11.12NLVTSPTKK214 pKa = 9.76IVPVKK219 pKa = 10.39EE220 pKa = 4.19EE221 pKa = 3.81EE222 pKa = 4.38GRR224 pKa = 11.84PAWAVKK230 pKa = 8.41QSRR233 pKa = 11.84KK234 pKa = 9.8LEE236 pKa = 4.08KK237 pKa = 10.13FRR239 pKa = 11.84QRR241 pKa = 11.84LNDD244 pKa = 3.25ANNRR248 pKa = 11.84QKK250 pKa = 11.23AGLPPKK256 pKa = 10.21RR257 pKa = 11.84KK258 pKa = 9.3PKK260 pKa = 9.96RR261 pKa = 11.84KK262 pKa = 9.33KK263 pKa = 10.17NLGAALPDD271 pKa = 3.56AYY273 pKa = 9.82KK274 pKa = 10.2EE275 pKa = 4.1RR276 pKa = 11.84KK277 pKa = 8.02RR278 pKa = 11.84QEE280 pKa = 3.75VEE282 pKa = 3.79RR283 pKa = 11.84KK284 pKa = 9.62RR285 pKa = 11.84KK286 pKa = 8.87EE287 pKa = 3.69KK288 pKa = 10.32RR289 pKa = 11.84KK290 pKa = 10.38KK291 pKa = 9.84ILLKK295 pKa = 10.81VGGRR299 pKa = 11.84SAA301 pKa = 4.04
MM1 pKa = 7.34NNSLTTQYY9 pKa = 11.32LGIPGEE15 pKa = 5.02DD16 pKa = 3.5PTSPPWASGNYY27 pKa = 9.33VGPFWSNGKK36 pKa = 8.68LQEE39 pKa = 4.24SVEE42 pKa = 4.39FGDD45 pKa = 5.04APPLHH50 pKa = 6.74EE51 pKa = 5.65LDD53 pKa = 3.66ALARR57 pKa = 11.84LHH59 pKa = 6.09DD60 pKa = 3.76TAYY63 pKa = 10.78ARR65 pKa = 11.84YY66 pKa = 9.22KK67 pKa = 10.92DD68 pKa = 3.75SAHH71 pKa = 6.8RR72 pKa = 11.84AAADD76 pKa = 3.37EE77 pKa = 4.36LFAEE81 pKa = 4.78EE82 pKa = 5.04AEE84 pKa = 4.32KK85 pKa = 10.89LKK87 pKa = 10.89KK88 pKa = 10.35KK89 pKa = 10.68YY90 pKa = 10.15GPNWAGNPQVAAKK103 pKa = 9.91LVRR106 pKa = 11.84YY107 pKa = 9.28GNHH110 pKa = 6.51TIRR113 pKa = 11.84AASRR117 pKa = 11.84VGGNVATGFKK127 pKa = 10.61FGGPLGAVGGLLYY140 pKa = 10.92SGLQNIKK147 pKa = 9.87QSHH150 pKa = 6.48DD151 pKa = 3.62MVTGNYY157 pKa = 9.02LRR159 pKa = 11.84KK160 pKa = 9.37EE161 pKa = 4.12RR162 pKa = 11.84KK163 pKa = 9.52DD164 pKa = 3.55LLSLFSKK171 pKa = 10.99DD172 pKa = 3.07PMKK175 pKa = 10.28TIKK178 pKa = 9.94VQPIGGSVVEE188 pKa = 4.93PGSSDD193 pKa = 3.42SKK195 pKa = 10.37PKK197 pKa = 10.64KK198 pKa = 8.52KK199 pKa = 8.95TLVEE203 pKa = 3.94KK204 pKa = 10.78LKK206 pKa = 11.12NLVTSPTKK214 pKa = 9.76IVPVKK219 pKa = 10.39EE220 pKa = 4.19EE221 pKa = 3.81EE222 pKa = 4.38GRR224 pKa = 11.84PAWAVKK230 pKa = 8.41QSRR233 pKa = 11.84KK234 pKa = 9.8LEE236 pKa = 4.08KK237 pKa = 10.13FRR239 pKa = 11.84QRR241 pKa = 11.84LNDD244 pKa = 3.25ANNRR248 pKa = 11.84QKK250 pKa = 11.23AGLPPKK256 pKa = 10.21RR257 pKa = 11.84KK258 pKa = 9.3PKK260 pKa = 9.96RR261 pKa = 11.84KK262 pKa = 9.33KK263 pKa = 10.17NLGAALPDD271 pKa = 3.56AYY273 pKa = 9.82KK274 pKa = 10.2EE275 pKa = 4.1RR276 pKa = 11.84KK277 pKa = 8.02RR278 pKa = 11.84QEE280 pKa = 3.75VEE282 pKa = 3.79RR283 pKa = 11.84KK284 pKa = 9.62RR285 pKa = 11.84KK286 pKa = 8.87EE287 pKa = 3.69KK288 pKa = 10.32RR289 pKa = 11.84KK290 pKa = 10.38KK291 pKa = 9.84ILLKK295 pKa = 10.81VGGRR299 pKa = 11.84SAA301 pKa = 4.04
Molecular weight: 33.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1225 |
301 |
548 |
408.3 |
45.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.755 ± 0.905 | 0.898 ± 0.287 |
4.98 ± 0.456 | 5.714 ± 1.418 |
3.673 ± 0.566 | 6.939 ± 0.657 |
2.531 ± 0.889 | 4.327 ± 0.636 |
7.673 ± 1.981 | 7.592 ± 0.647 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.122 ± 0.386 | 4.327 ± 0.865 |
6.122 ± 0.267 | 3.429 ± 0.119 |
5.469 ± 0.551 | 8.163 ± 1.069 |
6.041 ± 1.835 | 6.857 ± 0.274 |
1.551 ± 0.071 | 3.837 ± 0.314 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
