
Wuhan aphid virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
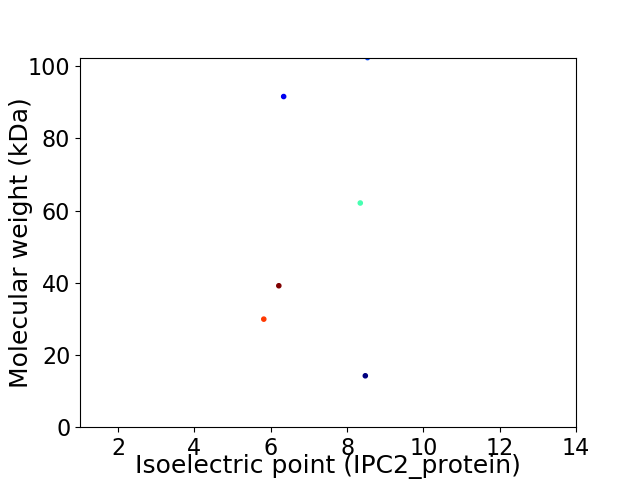
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0QL38|A0A0P0QL38_9VIRU Uncharacterized protein OS=Wuhan aphid virus 1 OX=1746067 GN=VP4 PE=4 SV=1
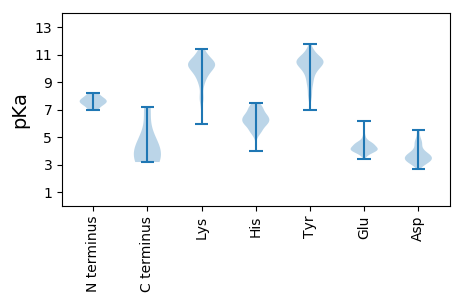
MM1 pKa = 7.64IAQGLLSLMLTATLATSLRR20 pKa = 11.84DD21 pKa = 3.5YY22 pKa = 10.9MRR24 pKa = 11.84DD25 pKa = 3.16EE26 pKa = 4.21NTLVICSEE34 pKa = 4.28YY35 pKa = 10.69SEE37 pKa = 4.29PWRR40 pKa = 11.84ISMCASSQLKK50 pKa = 10.64DD51 pKa = 3.68EE52 pKa = 5.54LISTHH57 pKa = 6.2TYY59 pKa = 7.74ITYY62 pKa = 9.98LQDD65 pKa = 4.77AMALLPGHH73 pKa = 6.28SKK75 pKa = 8.77YY76 pKa = 9.54TSIGVTTGHH85 pKa = 7.47DD86 pKa = 3.21KK87 pKa = 10.98RR88 pKa = 11.84IVLGQQLHH96 pKa = 6.01AHH98 pKa = 6.23FNWAYY103 pKa = 8.08EE104 pKa = 4.12AKK106 pKa = 9.93YY107 pKa = 10.53DD108 pKa = 3.91PRR110 pKa = 11.84DD111 pKa = 3.55YY112 pKa = 10.89YY113 pKa = 10.68TDD115 pKa = 3.38KK116 pKa = 10.85TFLAMRR122 pKa = 11.84YY123 pKa = 7.24KK124 pKa = 9.75TEE126 pKa = 3.98RR127 pKa = 11.84PILLFTVNYY136 pKa = 9.18EE137 pKa = 4.14PEE139 pKa = 4.0VPEE142 pKa = 4.08WNEE145 pKa = 3.87KK146 pKa = 8.86YY147 pKa = 10.51ISMLQPGGEE156 pKa = 3.94KK157 pKa = 10.26GAPRR161 pKa = 11.84LYY163 pKa = 9.91TGKK166 pKa = 10.59GYY168 pKa = 10.47RR169 pKa = 11.84ALTGKK174 pKa = 7.75QTEE177 pKa = 4.67DD178 pKa = 3.29CTSFYY183 pKa = 11.3DD184 pKa = 3.21WDD186 pKa = 3.59QRR188 pKa = 11.84EE189 pKa = 4.37SYY191 pKa = 8.09EE192 pKa = 4.28TAKK195 pKa = 9.44NTAILMDD202 pKa = 4.51PKK204 pKa = 10.37LWCPIIKK211 pKa = 10.15ASFEE215 pKa = 4.0IVVAEE220 pKa = 4.22IRR222 pKa = 11.84KK223 pKa = 5.91TTINTPHH230 pKa = 6.57GKK232 pKa = 7.7EE233 pKa = 4.09TIYY236 pKa = 9.91EE237 pKa = 4.55TVWEE241 pKa = 4.23NAGGRR246 pKa = 11.84LFGEE250 pKa = 4.85VPKK253 pKa = 10.56KK254 pKa = 9.72WSTVFLL260 pKa = 4.35
MM1 pKa = 7.64IAQGLLSLMLTATLATSLRR20 pKa = 11.84DD21 pKa = 3.5YY22 pKa = 10.9MRR24 pKa = 11.84DD25 pKa = 3.16EE26 pKa = 4.21NTLVICSEE34 pKa = 4.28YY35 pKa = 10.69SEE37 pKa = 4.29PWRR40 pKa = 11.84ISMCASSQLKK50 pKa = 10.64DD51 pKa = 3.68EE52 pKa = 5.54LISTHH57 pKa = 6.2TYY59 pKa = 7.74ITYY62 pKa = 9.98LQDD65 pKa = 4.77AMALLPGHH73 pKa = 6.28SKK75 pKa = 8.77YY76 pKa = 9.54TSIGVTTGHH85 pKa = 7.47DD86 pKa = 3.21KK87 pKa = 10.98RR88 pKa = 11.84IVLGQQLHH96 pKa = 6.01AHH98 pKa = 6.23FNWAYY103 pKa = 8.08EE104 pKa = 4.12AKK106 pKa = 9.93YY107 pKa = 10.53DD108 pKa = 3.91PRR110 pKa = 11.84DD111 pKa = 3.55YY112 pKa = 10.89YY113 pKa = 10.68TDD115 pKa = 3.38KK116 pKa = 10.85TFLAMRR122 pKa = 11.84YY123 pKa = 7.24KK124 pKa = 9.75TEE126 pKa = 3.98RR127 pKa = 11.84PILLFTVNYY136 pKa = 9.18EE137 pKa = 4.14PEE139 pKa = 4.0VPEE142 pKa = 4.08WNEE145 pKa = 3.87KK146 pKa = 8.86YY147 pKa = 10.51ISMLQPGGEE156 pKa = 3.94KK157 pKa = 10.26GAPRR161 pKa = 11.84LYY163 pKa = 9.91TGKK166 pKa = 10.59GYY168 pKa = 10.47RR169 pKa = 11.84ALTGKK174 pKa = 7.75QTEE177 pKa = 4.67DD178 pKa = 3.29CTSFYY183 pKa = 11.3DD184 pKa = 3.21WDD186 pKa = 3.59QRR188 pKa = 11.84EE189 pKa = 4.37SYY191 pKa = 8.09EE192 pKa = 4.28TAKK195 pKa = 9.44NTAILMDD202 pKa = 4.51PKK204 pKa = 10.37LWCPIIKK211 pKa = 10.15ASFEE215 pKa = 4.0IVVAEE220 pKa = 4.22IRR222 pKa = 11.84KK223 pKa = 5.91TTINTPHH230 pKa = 6.57GKK232 pKa = 7.7EE233 pKa = 4.09TIYY236 pKa = 9.91EE237 pKa = 4.55TVWEE241 pKa = 4.23NAGGRR246 pKa = 11.84LFGEE250 pKa = 4.85VPKK253 pKa = 10.56KK254 pKa = 9.72WSTVFLL260 pKa = 4.35
Molecular weight: 29.93 kDa
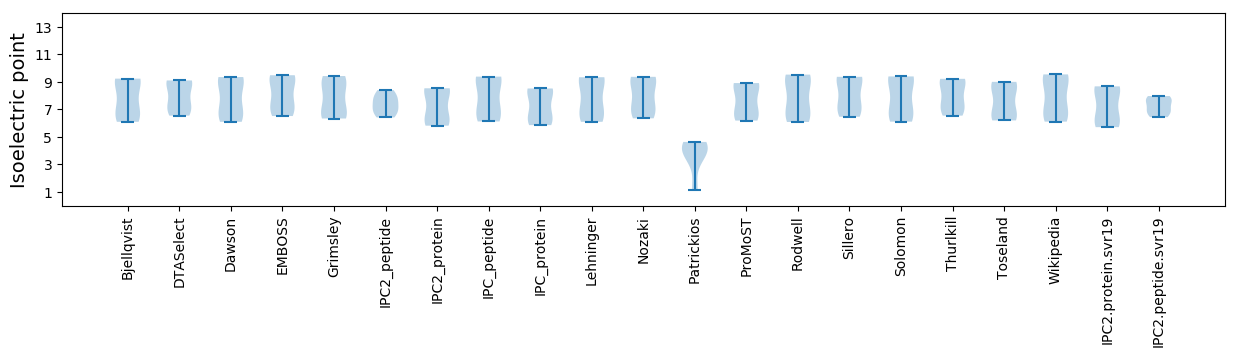
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0QL38|A0A0P0QL38_9VIRU Uncharacterized protein OS=Wuhan aphid virus 1 OX=1746067 GN=VP4 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.37ALYY5 pKa = 10.68LNAFCLFLIVCSTNAAQTTAAMNDD29 pKa = 3.93LEE31 pKa = 4.64SAWKK35 pKa = 9.92FDD37 pKa = 3.42LHH39 pKa = 6.12GFSVNPEE46 pKa = 3.7RR47 pKa = 11.84WDD49 pKa = 3.37DD50 pKa = 3.67ALRR53 pKa = 11.84LRR55 pKa = 11.84MYY57 pKa = 10.6QVGTAAAYY65 pKa = 8.74AATLPCSLPHH75 pKa = 6.27TLMFALVMFGMVRR88 pKa = 11.84LSWLLVAASCLALHH102 pKa = 7.01GYY104 pKa = 8.32KK105 pKa = 10.26RR106 pKa = 11.84KK107 pKa = 9.78NRR109 pKa = 11.84VFLGIGLALAYY120 pKa = 10.32ALAFGTKK127 pKa = 10.55LII129 pKa = 4.5
MM1 pKa = 7.51KK2 pKa = 10.37ALYY5 pKa = 10.68LNAFCLFLIVCSTNAAQTTAAMNDD29 pKa = 3.93LEE31 pKa = 4.64SAWKK35 pKa = 9.92FDD37 pKa = 3.42LHH39 pKa = 6.12GFSVNPEE46 pKa = 3.7RR47 pKa = 11.84WDD49 pKa = 3.37DD50 pKa = 3.67ALRR53 pKa = 11.84LRR55 pKa = 11.84MYY57 pKa = 10.6QVGTAAAYY65 pKa = 8.74AATLPCSLPHH75 pKa = 6.27TLMFALVMFGMVRR88 pKa = 11.84LSWLLVAASCLALHH102 pKa = 7.01GYY104 pKa = 8.32KK105 pKa = 10.26RR106 pKa = 11.84KK107 pKa = 9.78NRR109 pKa = 11.84VFLGIGLALAYY120 pKa = 10.32ALAFGTKK127 pKa = 10.55LII129 pKa = 4.5
Molecular weight: 14.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2993 |
129 |
907 |
498.8 |
56.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.148 ± 0.857 | 1.437 ± 0.268 |
4.911 ± 0.501 | 4.945 ± 0.44 |
4.277 ± 0.229 | 7.016 ± 0.492 |
2.74 ± 0.354 | 6.682 ± 0.4 |
5.179 ± 0.392 | 10.057 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.408 ± 0.245 | 3.775 ± 0.367 |
4.611 ± 0.14 | 3.508 ± 0.222 |
5.446 ± 0.299 | 5.78 ± 0.252 |
7.584 ± 0.607 | 5.981 ± 0.294 |
1.871 ± 0.297 | 4.577 ± 0.436 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
