
Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003) (Inky cap fungus) (Hormographiella aspergillata)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
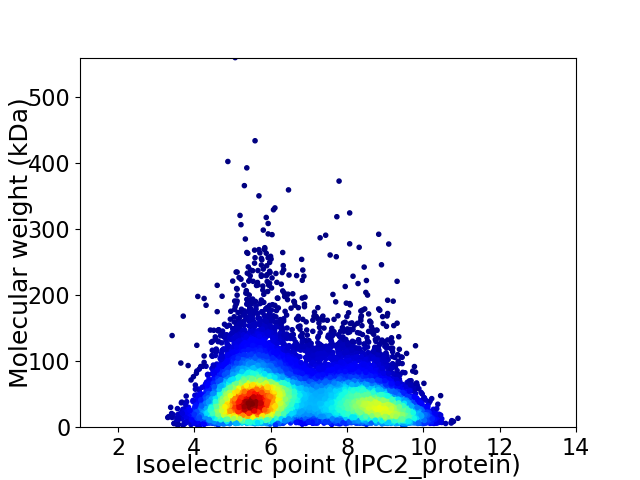
Virtual 2D-PAGE plot for 13335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8NHD5|A8NHD5_COPC7 Uncharacterized protein OS=Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003) OX=240176 GN=CC1G_11527 PE=4 SV=2
MM1 pKa = 6.81TTDD4 pKa = 3.14STSLFDD10 pKa = 4.15LARR13 pKa = 11.84NKK15 pKa = 10.5LQTSFGGHH23 pKa = 6.64KK24 pKa = 10.12DD25 pKa = 2.85RR26 pKa = 11.84CSLHH30 pKa = 6.24RR31 pKa = 11.84WVLLKK36 pKa = 10.99NSIINSTSALVSSAVDD52 pKa = 3.27AVSEE56 pKa = 4.46TQVSSYY62 pKa = 11.44SEE64 pKa = 5.02DD65 pKa = 4.37LDD67 pKa = 6.47DD68 pKa = 6.32DD69 pKa = 5.59DD70 pKa = 5.53SVSDD74 pKa = 3.82EE75 pKa = 4.33VLSSVAGGSVHH86 pKa = 7.63SFMFPDD92 pKa = 3.97AGNLVEE98 pKa = 5.7AGPADD103 pKa = 3.54AHH105 pKa = 6.73SSEE108 pKa = 4.27AQWLDD113 pKa = 3.3SLLEE117 pKa = 4.07ALGDD121 pKa = 4.95DD122 pKa = 5.06DD123 pKa = 7.11DD124 pKa = 7.39DD125 pKa = 6.35DD126 pKa = 5.38FGPDD130 pKa = 3.1SDD132 pKa = 5.44SRR134 pKa = 11.84TEE136 pKa = 5.39DD137 pKa = 3.13EE138 pKa = 5.26DD139 pKa = 3.68EE140 pKa = 4.31QLFSPSASPMSSSDD154 pKa = 3.68DD155 pKa = 4.03LLPSMHH161 pKa = 6.32NHH163 pKa = 6.64HH164 pKa = 7.48PFQQHH169 pKa = 3.47QHH171 pKa = 5.33HH172 pKa = 7.19SYY174 pKa = 10.54FSSLPVSYY182 pKa = 9.71PIIDD186 pKa = 4.87PYY188 pKa = 10.31PPCHH192 pKa = 6.66PPLASPYY199 pKa = 10.42AFDD202 pKa = 4.1SSSLSSLPPPYY213 pKa = 10.07EE214 pKa = 4.67DD215 pKa = 4.72PLPFSDD221 pKa = 5.93DD222 pKa = 3.45VDD224 pKa = 4.25DD225 pKa = 4.67MPVPDD230 pKa = 4.95TIEE233 pKa = 5.43DD234 pKa = 3.61NTSDD238 pKa = 4.94DD239 pKa = 4.48EE240 pKa = 4.96SDD242 pKa = 4.02SPPTPMGGSRR252 pKa = 11.84SSLFDD257 pKa = 3.42VDD259 pKa = 4.32VLDD262 pKa = 4.62AASIPLPPDD271 pKa = 3.34RR272 pKa = 11.84SRR274 pKa = 11.84LSHH277 pKa = 5.89GAAVRR282 pKa = 11.84VYY284 pKa = 10.14DD285 pKa = 3.9QDD287 pKa = 4.03SSSSSCFGPSFDD299 pKa = 4.92PLPFSTNDD307 pKa = 2.93VHH309 pKa = 6.57STYY312 pKa = 11.27NLYY315 pKa = 10.57SEE317 pKa = 4.91CC318 pKa = 4.87
MM1 pKa = 6.81TTDD4 pKa = 3.14STSLFDD10 pKa = 4.15LARR13 pKa = 11.84NKK15 pKa = 10.5LQTSFGGHH23 pKa = 6.64KK24 pKa = 10.12DD25 pKa = 2.85RR26 pKa = 11.84CSLHH30 pKa = 6.24RR31 pKa = 11.84WVLLKK36 pKa = 10.99NSIINSTSALVSSAVDD52 pKa = 3.27AVSEE56 pKa = 4.46TQVSSYY62 pKa = 11.44SEE64 pKa = 5.02DD65 pKa = 4.37LDD67 pKa = 6.47DD68 pKa = 6.32DD69 pKa = 5.59DD70 pKa = 5.53SVSDD74 pKa = 3.82EE75 pKa = 4.33VLSSVAGGSVHH86 pKa = 7.63SFMFPDD92 pKa = 3.97AGNLVEE98 pKa = 5.7AGPADD103 pKa = 3.54AHH105 pKa = 6.73SSEE108 pKa = 4.27AQWLDD113 pKa = 3.3SLLEE117 pKa = 4.07ALGDD121 pKa = 4.95DD122 pKa = 5.06DD123 pKa = 7.11DD124 pKa = 7.39DD125 pKa = 6.35DD126 pKa = 5.38FGPDD130 pKa = 3.1SDD132 pKa = 5.44SRR134 pKa = 11.84TEE136 pKa = 5.39DD137 pKa = 3.13EE138 pKa = 5.26DD139 pKa = 3.68EE140 pKa = 4.31QLFSPSASPMSSSDD154 pKa = 3.68DD155 pKa = 4.03LLPSMHH161 pKa = 6.32NHH163 pKa = 6.64HH164 pKa = 7.48PFQQHH169 pKa = 3.47QHH171 pKa = 5.33HH172 pKa = 7.19SYY174 pKa = 10.54FSSLPVSYY182 pKa = 9.71PIIDD186 pKa = 4.87PYY188 pKa = 10.31PPCHH192 pKa = 6.66PPLASPYY199 pKa = 10.42AFDD202 pKa = 4.1SSSLSSLPPPYY213 pKa = 10.07EE214 pKa = 4.67DD215 pKa = 4.72PLPFSDD221 pKa = 5.93DD222 pKa = 3.45VDD224 pKa = 4.25DD225 pKa = 4.67MPVPDD230 pKa = 4.95TIEE233 pKa = 5.43DD234 pKa = 3.61NTSDD238 pKa = 4.94DD239 pKa = 4.48EE240 pKa = 4.96SDD242 pKa = 4.02SPPTPMGGSRR252 pKa = 11.84SSLFDD257 pKa = 3.42VDD259 pKa = 4.32VLDD262 pKa = 4.62AASIPLPPDD271 pKa = 3.34RR272 pKa = 11.84SRR274 pKa = 11.84LSHH277 pKa = 5.89GAAVRR282 pKa = 11.84VYY284 pKa = 10.14DD285 pKa = 3.9QDD287 pKa = 4.03SSSSSCFGPSFDD299 pKa = 4.92PLPFSTNDD307 pKa = 2.93VHH309 pKa = 6.57STYY312 pKa = 11.27NLYY315 pKa = 10.57SEE317 pKa = 4.91CC318 pKa = 4.87
Molecular weight: 34.38 kDa
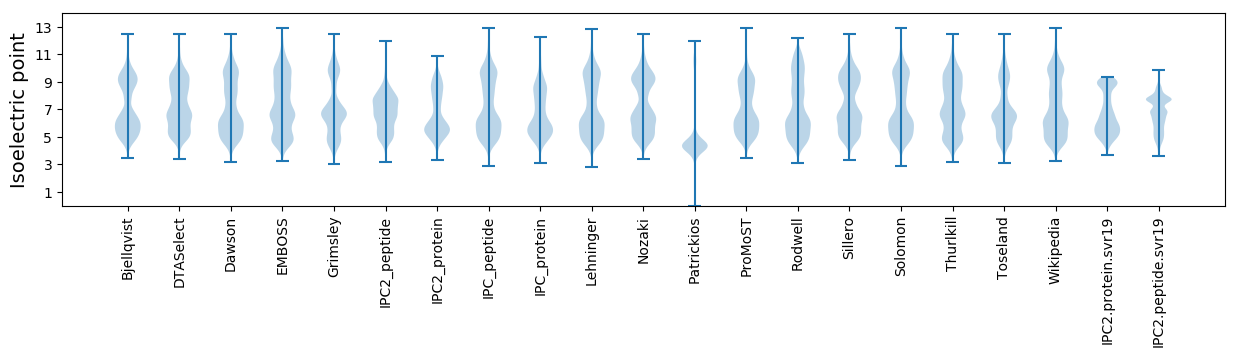
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6RQP7|D6RQP7_COPC7 Uncharacterized protein OS=Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003) OX=240176 GN=CC1G_15455 PE=4 SV=1
MM1 pKa = 7.56AVLDD5 pKa = 3.9VFGLVLKK12 pKa = 10.41GGRR15 pKa = 11.84RR16 pKa = 11.84VRR18 pKa = 11.84QPLTRR23 pKa = 11.84TRR25 pKa = 11.84TRR27 pKa = 11.84SLPRR31 pKa = 11.84TRR33 pKa = 11.84NPPGIEE39 pKa = 4.13NPCPSLDD46 pKa = 3.68VFSSLSGRR54 pKa = 11.84WGAQAEE60 pKa = 4.38RR61 pKa = 11.84CNDD64 pKa = 3.01SHH66 pKa = 7.25RR67 pKa = 11.84KK68 pKa = 9.01KK69 pKa = 10.03NHH71 pKa = 6.05PSAAYY76 pKa = 10.48SFFDD80 pKa = 3.86HH81 pKa = 6.87PTNSLRR87 pKa = 11.84QRR89 pKa = 11.84AWLVFSVGGLVRR101 pKa = 11.84YY102 pKa = 8.58AALRR106 pKa = 11.84KK107 pKa = 8.73VTMNARR113 pKa = 11.84GALASFTSGRR123 pKa = 11.84EE124 pKa = 3.85FGNWWCQGRR133 pKa = 11.84SPVDD137 pKa = 3.04AFAFILPKK145 pKa = 10.38MMVPSLKK152 pKa = 9.86TLVLEE157 pKa = 4.5TRR159 pKa = 11.84YY160 pKa = 9.98HH161 pKa = 6.26RR162 pKa = 11.84PALFAAIVHH171 pKa = 6.55EE172 pKa = 4.67DD173 pKa = 3.92SDD175 pKa = 4.14WEE177 pKa = 4.14TLDD180 pKa = 4.29FVSCRR185 pKa = 11.84VTMVQDD191 pKa = 3.49
MM1 pKa = 7.56AVLDD5 pKa = 3.9VFGLVLKK12 pKa = 10.41GGRR15 pKa = 11.84RR16 pKa = 11.84VRR18 pKa = 11.84QPLTRR23 pKa = 11.84TRR25 pKa = 11.84TRR27 pKa = 11.84SLPRR31 pKa = 11.84TRR33 pKa = 11.84NPPGIEE39 pKa = 4.13NPCPSLDD46 pKa = 3.68VFSSLSGRR54 pKa = 11.84WGAQAEE60 pKa = 4.38RR61 pKa = 11.84CNDD64 pKa = 3.01SHH66 pKa = 7.25RR67 pKa = 11.84KK68 pKa = 9.01KK69 pKa = 10.03NHH71 pKa = 6.05PSAAYY76 pKa = 10.48SFFDD80 pKa = 3.86HH81 pKa = 6.87PTNSLRR87 pKa = 11.84QRR89 pKa = 11.84AWLVFSVGGLVRR101 pKa = 11.84YY102 pKa = 8.58AALRR106 pKa = 11.84KK107 pKa = 8.73VTMNARR113 pKa = 11.84GALASFTSGRR123 pKa = 11.84EE124 pKa = 3.85FGNWWCQGRR133 pKa = 11.84SPVDD137 pKa = 3.04AFAFILPKK145 pKa = 10.38MMVPSLKK152 pKa = 9.86TLVLEE157 pKa = 4.5TRR159 pKa = 11.84YY160 pKa = 9.98HH161 pKa = 6.26RR162 pKa = 11.84PALFAAIVHH171 pKa = 6.55EE172 pKa = 4.67DD173 pKa = 3.92SDD175 pKa = 4.14WEE177 pKa = 4.14TLDD180 pKa = 4.29FVSCRR185 pKa = 11.84VTMVQDD191 pKa = 3.49
Molecular weight: 21.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6160588 |
27 |
4998 |
462.0 |
51.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.129 ± 0.019 | 1.164 ± 0.009 |
5.653 ± 0.014 | 6.215 ± 0.022 |
3.654 ± 0.017 | 6.654 ± 0.022 |
2.493 ± 0.01 | 4.633 ± 0.018 |
4.76 ± 0.018 | 9.042 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.927 ± 0.008 | 3.517 ± 0.012 |
6.922 ± 0.029 | 3.767 ± 0.015 |
6.402 ± 0.02 | 8.745 ± 0.029 |
6.035 ± 0.013 | 6.232 ± 0.016 |
1.42 ± 0.009 | 2.636 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
