
Trypanosoma cruzi Dm28c
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Trypanosoma; Schizotrypanum; Trypanosoma cruzi
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
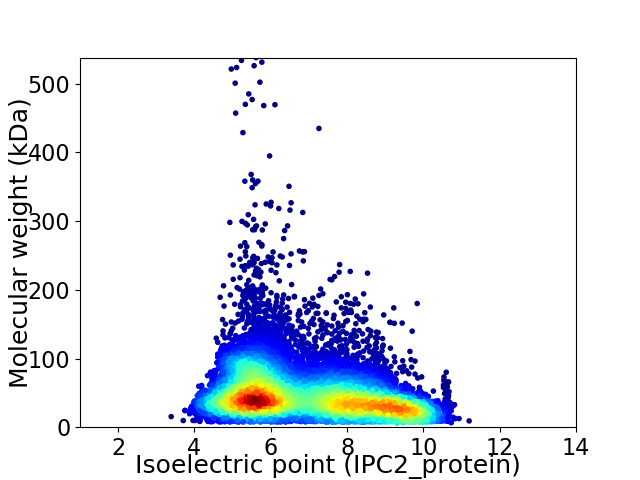
Virtual 2D-PAGE plot for 11346 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5B519|V5B519_TRYCR Uncharacterized protein OS=Trypanosoma cruzi Dm28c OX=1416333 GN=TCDM_12245 PE=4 SV=1
MM1 pKa = 6.98MCEE4 pKa = 3.85EE5 pKa = 5.26CFVDD9 pKa = 3.23WWNSKK14 pKa = 8.93IYY16 pKa = 10.06IYY18 pKa = 10.26IFCEE22 pKa = 3.46VHH24 pKa = 5.73YY25 pKa = 10.05LYY27 pKa = 7.78TTSCFYY33 pKa = 10.99FPACNMKK40 pKa = 10.66LKK42 pKa = 10.21FFWIDD47 pKa = 3.23FSLFFWDD54 pKa = 4.62FCRR57 pKa = 11.84SVNFLFFFLVAFIGLMFLMKK77 pKa = 10.56CMLLPLVFFTAWLCDD92 pKa = 3.46LSLFLLSLCVDD103 pKa = 3.75VLLVCAEE110 pKa = 4.1GCTQVTGVMAMMMTGRR126 pKa = 11.84VLLVCALCVLWCGAGGRR143 pKa = 11.84CDD145 pKa = 3.8EE146 pKa = 5.03GEE148 pKa = 4.11DD149 pKa = 3.78TAGSGSGGEE158 pKa = 4.2HH159 pKa = 6.32SLASKK164 pKa = 10.39QLEE167 pKa = 4.15NSSQDD172 pKa = 3.49TQSLKK177 pKa = 11.04GGVPGAKK184 pKa = 10.03GNVPPAPTDD193 pKa = 3.64EE194 pKa = 4.97EE195 pKa = 6.34DD196 pKa = 4.63DD197 pKa = 5.83DD198 pKa = 4.93SDD200 pKa = 4.02NDD202 pKa = 3.5EE203 pKa = 5.02DD204 pKa = 3.95EE205 pKa = 4.41GTEE208 pKa = 4.14VEE210 pKa = 4.53EE211 pKa = 4.32KK212 pKa = 10.48TIGRR216 pKa = 11.84QGGQGGTTVPGPDD229 pKa = 3.2STEE232 pKa = 3.86TNLIGTEE239 pKa = 4.02QQTGQSIISAEE250 pKa = 4.36EE251 pKa = 3.79IPPSGSQEE259 pKa = 3.79PKK261 pKa = 10.77VILTQPEE268 pKa = 4.27VEE270 pKa = 4.71GKK272 pKa = 10.24NDD274 pKa = 3.6TDD276 pKa = 3.4KK277 pKa = 11.44SRR279 pKa = 11.84PAVEE283 pKa = 4.21NALTTVNGEE292 pKa = 3.81NTLPGGVNLPSPPEE306 pKa = 4.46DD307 pKa = 3.4GVDD310 pKa = 3.2SHH312 pKa = 6.12EE313 pKa = 4.39QSGEE317 pKa = 3.95DD318 pKa = 3.3TTSEE322 pKa = 4.09DD323 pKa = 3.84EE324 pKa = 4.59KK325 pKa = 11.16NVPSPEE331 pKa = 3.97TAATPQSHH339 pKa = 6.56RR340 pKa = 11.84GEE342 pKa = 4.4GSEE345 pKa = 4.57GTGEE349 pKa = 4.0DD350 pKa = 3.46TKK352 pKa = 11.26ATTLTANTTDD362 pKa = 3.32TTNTQNSDD370 pKa = 3.0SSTVKK375 pKa = 10.14MNEE378 pKa = 3.76AAPQTTITLTAAQTNHH394 pKa = 5.27TVTTGNSDD402 pKa = 2.8GSTAVSHH409 pKa = 5.47TTSSLLLLLLFACAAAAAVVAAA431 pKa = 4.95
MM1 pKa = 6.98MCEE4 pKa = 3.85EE5 pKa = 5.26CFVDD9 pKa = 3.23WWNSKK14 pKa = 8.93IYY16 pKa = 10.06IYY18 pKa = 10.26IFCEE22 pKa = 3.46VHH24 pKa = 5.73YY25 pKa = 10.05LYY27 pKa = 7.78TTSCFYY33 pKa = 10.99FPACNMKK40 pKa = 10.66LKK42 pKa = 10.21FFWIDD47 pKa = 3.23FSLFFWDD54 pKa = 4.62FCRR57 pKa = 11.84SVNFLFFFLVAFIGLMFLMKK77 pKa = 10.56CMLLPLVFFTAWLCDD92 pKa = 3.46LSLFLLSLCVDD103 pKa = 3.75VLLVCAEE110 pKa = 4.1GCTQVTGVMAMMMTGRR126 pKa = 11.84VLLVCALCVLWCGAGGRR143 pKa = 11.84CDD145 pKa = 3.8EE146 pKa = 5.03GEE148 pKa = 4.11DD149 pKa = 3.78TAGSGSGGEE158 pKa = 4.2HH159 pKa = 6.32SLASKK164 pKa = 10.39QLEE167 pKa = 4.15NSSQDD172 pKa = 3.49TQSLKK177 pKa = 11.04GGVPGAKK184 pKa = 10.03GNVPPAPTDD193 pKa = 3.64EE194 pKa = 4.97EE195 pKa = 6.34DD196 pKa = 4.63DD197 pKa = 5.83DD198 pKa = 4.93SDD200 pKa = 4.02NDD202 pKa = 3.5EE203 pKa = 5.02DD204 pKa = 3.95EE205 pKa = 4.41GTEE208 pKa = 4.14VEE210 pKa = 4.53EE211 pKa = 4.32KK212 pKa = 10.48TIGRR216 pKa = 11.84QGGQGGTTVPGPDD229 pKa = 3.2STEE232 pKa = 3.86TNLIGTEE239 pKa = 4.02QQTGQSIISAEE250 pKa = 4.36EE251 pKa = 3.79IPPSGSQEE259 pKa = 3.79PKK261 pKa = 10.77VILTQPEE268 pKa = 4.27VEE270 pKa = 4.71GKK272 pKa = 10.24NDD274 pKa = 3.6TDD276 pKa = 3.4KK277 pKa = 11.44SRR279 pKa = 11.84PAVEE283 pKa = 4.21NALTTVNGEE292 pKa = 3.81NTLPGGVNLPSPPEE306 pKa = 4.46DD307 pKa = 3.4GVDD310 pKa = 3.2SHH312 pKa = 6.12EE313 pKa = 4.39QSGEE317 pKa = 3.95DD318 pKa = 3.3TTSEE322 pKa = 4.09DD323 pKa = 3.84EE324 pKa = 4.59KK325 pKa = 11.16NVPSPEE331 pKa = 3.97TAATPQSHH339 pKa = 6.56RR340 pKa = 11.84GEE342 pKa = 4.4GSEE345 pKa = 4.57GTGEE349 pKa = 4.0DD350 pKa = 3.46TKK352 pKa = 11.26ATTLTANTTDD362 pKa = 3.32TTNTQNSDD370 pKa = 3.0SSTVKK375 pKa = 10.14MNEE378 pKa = 3.76AAPQTTITLTAAQTNHH394 pKa = 5.27TVTTGNSDD402 pKa = 2.8GSTAVSHH409 pKa = 5.47TTSSLLLLLLFACAAAAAVVAAA431 pKa = 4.95
Molecular weight: 45.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5AN66|V5AN66_TRYCR Uncharacterized protein OS=Trypanosoma cruzi Dm28c OX=1416333 GN=TCDM_13482 PE=4 SV=1
MM1 pKa = 7.4HH2 pKa = 7.41KK3 pKa = 10.12KK4 pKa = 9.87KK5 pKa = 10.19KK6 pKa = 9.58RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.41RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57KK14 pKa = 10.33RR15 pKa = 11.84NNKK18 pKa = 9.66RR19 pKa = 11.84ILRR22 pKa = 11.84MKK24 pKa = 10.07TKK26 pKa = 10.53TNTKK30 pKa = 9.39KK31 pKa = 10.69RR32 pKa = 11.84KK33 pKa = 6.05TKK35 pKa = 9.37KK36 pKa = 9.5RR37 pKa = 11.84KK38 pKa = 9.28RR39 pKa = 11.84KK40 pKa = 9.4RR41 pKa = 11.84KK42 pKa = 8.36RR43 pKa = 11.84KK44 pKa = 8.12GMKK47 pKa = 9.65RR48 pKa = 11.84KK49 pKa = 7.56TEE51 pKa = 3.72GMVRR55 pKa = 11.84MKK57 pKa = 10.69RR58 pKa = 11.84GIQRR62 pKa = 11.84KK63 pKa = 9.01KK64 pKa = 10.6KK65 pKa = 10.3LLPTPQRR72 pKa = 3.52
MM1 pKa = 7.4HH2 pKa = 7.41KK3 pKa = 10.12KK4 pKa = 9.87KK5 pKa = 10.19KK6 pKa = 9.58RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.41RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57KK14 pKa = 10.33RR15 pKa = 11.84NNKK18 pKa = 9.66RR19 pKa = 11.84ILRR22 pKa = 11.84MKK24 pKa = 10.07TKK26 pKa = 10.53TNTKK30 pKa = 9.39KK31 pKa = 10.69RR32 pKa = 11.84KK33 pKa = 6.05TKK35 pKa = 9.37KK36 pKa = 9.5RR37 pKa = 11.84KK38 pKa = 9.28RR39 pKa = 11.84KK40 pKa = 9.4RR41 pKa = 11.84KK42 pKa = 8.36RR43 pKa = 11.84KK44 pKa = 8.12GMKK47 pKa = 9.65RR48 pKa = 11.84KK49 pKa = 7.56TEE51 pKa = 3.72GMVRR55 pKa = 11.84MKK57 pKa = 10.69RR58 pKa = 11.84GIQRR62 pKa = 11.84KK63 pKa = 9.01KK64 pKa = 10.6KK65 pKa = 10.3LLPTPQRR72 pKa = 3.52
Molecular weight: 9.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5280116 |
51 |
4838 |
465.4 |
51.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.477 ± 0.024 | 2.255 ± 0.014 |
4.732 ± 0.013 | 6.823 ± 0.029 |
3.736 ± 0.015 | 6.73 ± 0.026 |
2.677 ± 0.011 | 3.792 ± 0.014 |
4.416 ± 0.022 | 9.562 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.007 | 3.542 ± 0.011 |
4.965 ± 0.018 | 3.745 ± 0.018 |
7.083 ± 0.023 | 8.009 ± 0.025 |
5.872 ± 0.016 | 7.476 ± 0.028 |
1.249 ± 0.007 | 2.326 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
