
Sewage-associated circular DNA virus-19
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
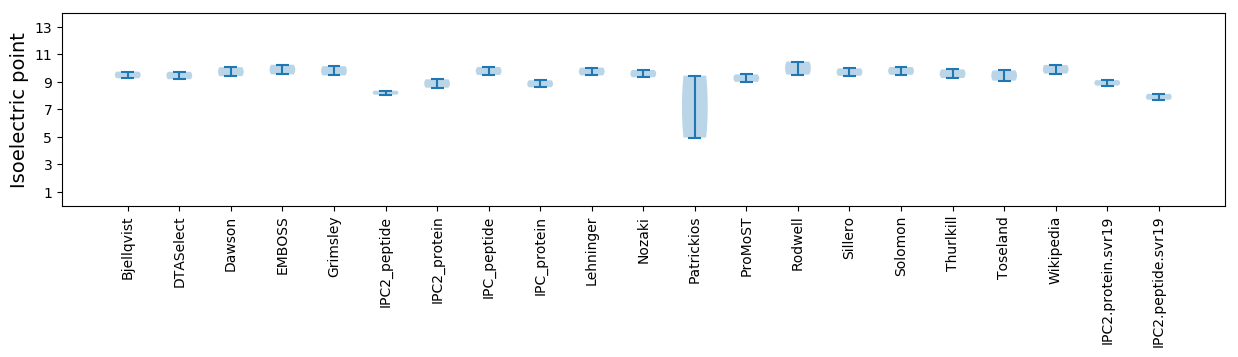
Average proteome isoelectric point is 8.93
Get precalculated fractions of proteins
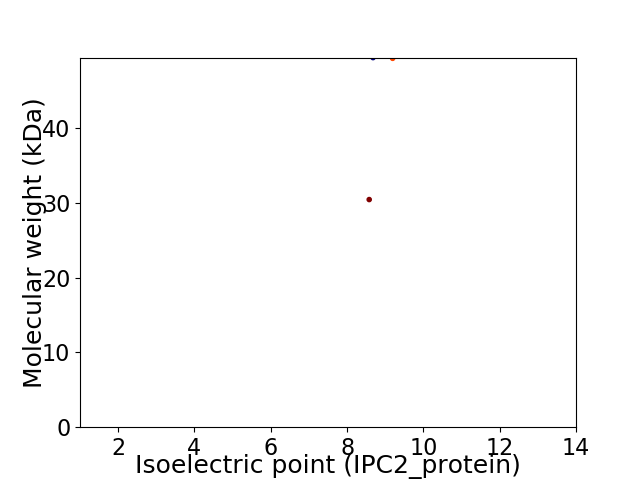
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI41|A0A0B4UI41_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-19 OX=1592086 PE=4 SV=1
MM1 pKa = 6.8SQARR5 pKa = 11.84FWILTIPHH13 pKa = 7.26ADD15 pKa = 3.51FTPYY19 pKa = 10.68LPPNVNHH26 pKa = 7.34ISGQLEE32 pKa = 4.1RR33 pKa = 11.84GTGLNQYY40 pKa = 7.37LHH42 pKa = 5.49WQIVVSFARR51 pKa = 11.84KK52 pKa = 8.88IRR54 pKa = 11.84LGGVKK59 pKa = 10.2SIFGDD64 pKa = 3.57SCHH67 pKa = 7.09AEE69 pKa = 4.11PTRR72 pKa = 11.84SDD74 pKa = 3.12AARR77 pKa = 11.84KK78 pKa = 8.88YY79 pKa = 7.54VHH81 pKa = 6.93KK82 pKa = 10.57KK83 pKa = 7.57DD84 pKa = 3.5TRR86 pKa = 11.84VDD88 pKa = 3.38GTGFEE93 pKa = 5.19LGTLPIRR100 pKa = 11.84RR101 pKa = 11.84GEE103 pKa = 4.2SCDD106 pKa = 3.2WEE108 pKa = 4.25AVRR111 pKa = 11.84EE112 pKa = 4.0NAKK115 pKa = 10.1RR116 pKa = 11.84GRR118 pKa = 11.84MDD120 pKa = 6.55DD121 pKa = 3.18IPADD125 pKa = 3.69IFCRR129 pKa = 11.84LYY131 pKa = 11.52GNLKK135 pKa = 10.23RR136 pKa = 11.84IAVDD140 pKa = 3.15NMAPIGLEE148 pKa = 3.72RR149 pKa = 11.84TINVYY154 pKa = 9.07WGRR157 pKa = 11.84TGTGKK162 pKa = 10.01SRR164 pKa = 11.84RR165 pKa = 11.84AWAEE169 pKa = 3.3AGMDD173 pKa = 4.61AFPKK177 pKa = 10.51DD178 pKa = 3.77PRR180 pKa = 11.84TKK182 pKa = 9.94FWDD185 pKa = 4.39GYY187 pKa = 8.38RR188 pKa = 11.84SHH190 pKa = 6.73RR191 pKa = 11.84HH192 pKa = 3.57VVIDD196 pKa = 3.79EE197 pKa = 4.0FRR199 pKa = 11.84GGIDD203 pKa = 3.25VAHH206 pKa = 7.59LLRR209 pKa = 11.84WFDD212 pKa = 3.74RR213 pKa = 11.84YY214 pKa = 10.05PVVVEE219 pKa = 4.03VKK221 pKa = 10.34GSSVVLAAEE230 pKa = 4.51TVWITSNLDD239 pKa = 3.01PRR241 pKa = 11.84QWYY244 pKa = 8.69PDD246 pKa = 3.61LDD248 pKa = 4.18EE249 pKa = 4.59EE250 pKa = 4.49TLAALLRR257 pKa = 11.84RR258 pKa = 11.84LNITHH263 pKa = 6.07FTII266 pKa = 4.86
MM1 pKa = 6.8SQARR5 pKa = 11.84FWILTIPHH13 pKa = 7.26ADD15 pKa = 3.51FTPYY19 pKa = 10.68LPPNVNHH26 pKa = 7.34ISGQLEE32 pKa = 4.1RR33 pKa = 11.84GTGLNQYY40 pKa = 7.37LHH42 pKa = 5.49WQIVVSFARR51 pKa = 11.84KK52 pKa = 8.88IRR54 pKa = 11.84LGGVKK59 pKa = 10.2SIFGDD64 pKa = 3.57SCHH67 pKa = 7.09AEE69 pKa = 4.11PTRR72 pKa = 11.84SDD74 pKa = 3.12AARR77 pKa = 11.84KK78 pKa = 8.88YY79 pKa = 7.54VHH81 pKa = 6.93KK82 pKa = 10.57KK83 pKa = 7.57DD84 pKa = 3.5TRR86 pKa = 11.84VDD88 pKa = 3.38GTGFEE93 pKa = 5.19LGTLPIRR100 pKa = 11.84RR101 pKa = 11.84GEE103 pKa = 4.2SCDD106 pKa = 3.2WEE108 pKa = 4.25AVRR111 pKa = 11.84EE112 pKa = 4.0NAKK115 pKa = 10.1RR116 pKa = 11.84GRR118 pKa = 11.84MDD120 pKa = 6.55DD121 pKa = 3.18IPADD125 pKa = 3.69IFCRR129 pKa = 11.84LYY131 pKa = 11.52GNLKK135 pKa = 10.23RR136 pKa = 11.84IAVDD140 pKa = 3.15NMAPIGLEE148 pKa = 3.72RR149 pKa = 11.84TINVYY154 pKa = 9.07WGRR157 pKa = 11.84TGTGKK162 pKa = 10.01SRR164 pKa = 11.84RR165 pKa = 11.84AWAEE169 pKa = 3.3AGMDD173 pKa = 4.61AFPKK177 pKa = 10.51DD178 pKa = 3.77PRR180 pKa = 11.84TKK182 pKa = 9.94FWDD185 pKa = 4.39GYY187 pKa = 8.38RR188 pKa = 11.84SHH190 pKa = 6.73RR191 pKa = 11.84HH192 pKa = 3.57VVIDD196 pKa = 3.79EE197 pKa = 4.0FRR199 pKa = 11.84GGIDD203 pKa = 3.25VAHH206 pKa = 7.59LLRR209 pKa = 11.84WFDD212 pKa = 3.74RR213 pKa = 11.84YY214 pKa = 10.05PVVVEE219 pKa = 4.03VKK221 pKa = 10.34GSSVVLAAEE230 pKa = 4.51TVWITSNLDD239 pKa = 3.01PRR241 pKa = 11.84QWYY244 pKa = 8.69PDD246 pKa = 3.61LDD248 pKa = 4.18EE249 pKa = 4.59EE250 pKa = 4.49TLAALLRR257 pKa = 11.84RR258 pKa = 11.84LNITHH263 pKa = 6.07FTII266 pKa = 4.86
Molecular weight: 30.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI41|A0A0B4UI41_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-19 OX=1592086 PE=4 SV=1
MM1 pKa = 7.16SVVRR5 pKa = 11.84KK6 pKa = 8.61RR7 pKa = 11.84TGGYY11 pKa = 5.71TTYY14 pKa = 11.3APNKK18 pKa = 9.1KK19 pKa = 8.59MRR21 pKa = 11.84SGRR24 pKa = 11.84DD25 pKa = 3.15ILVSAGLAASRR36 pKa = 11.84KK37 pKa = 8.04YY38 pKa = 10.69GPRR41 pKa = 11.84LVSMAAQKK49 pKa = 9.93LASMAYY55 pKa = 9.8AAAKK59 pKa = 9.89DD60 pKa = 3.77DD61 pKa = 5.53PITQFRR67 pKa = 11.84NAKK70 pKa = 9.47KK71 pKa = 8.16RR72 pKa = 11.84TKK74 pKa = 9.92GRR76 pKa = 11.84SYY78 pKa = 11.28SEE80 pKa = 4.1QKK82 pKa = 10.59GVAAGKK88 pKa = 9.75FRR90 pKa = 11.84ATKK93 pKa = 10.36RR94 pKa = 11.84SPKK97 pKa = 10.35NIWLKK102 pKa = 7.72YY103 pKa = 6.6TRR105 pKa = 11.84KK106 pKa = 10.31GCVTTKK112 pKa = 10.37EE113 pKa = 4.29HH114 pKa = 6.32GCVLTGTVNTNAIYY128 pKa = 10.18FGHH131 pKa = 5.78STHH134 pKa = 7.04ANTYY138 pKa = 10.89LMLTVYY144 pKa = 9.48MRR146 pKa = 11.84AIVIHH151 pKa = 6.6IFRR154 pKa = 11.84KK155 pKa = 10.23GGVQIEE161 pKa = 4.65DD162 pKa = 4.12IEE164 pKa = 4.61TEE166 pKa = 4.31SPGSGGSMTIEE177 pKa = 3.73YY178 pKa = 8.73TYY180 pKa = 10.4NTDD183 pKa = 2.76IANQFVTVAFTGGLTLNQIAGSFASQFTNILFNNANDD220 pKa = 3.68EE221 pKa = 4.18QRR223 pKa = 11.84FFPILEE229 pKa = 4.27AKK231 pKa = 10.21IEE233 pKa = 4.06IPNLEE238 pKa = 4.56LCKK241 pKa = 9.93QDD243 pKa = 3.32YY244 pKa = 10.26RR245 pKa = 11.84NSIVHH250 pKa = 5.94VMVKK254 pKa = 10.38SDD256 pKa = 4.09LKK258 pKa = 9.95VQNRR262 pKa = 11.84TLATGTDD269 pKa = 3.45EE270 pKa = 5.82DD271 pKa = 4.59NNSSEE276 pKa = 4.85NIANQPLYY284 pKa = 11.16GKK286 pKa = 10.05FYY288 pKa = 9.59HH289 pKa = 6.78GKK291 pKa = 9.65GNGVFARR298 pKa = 11.84ANAGAGTGKK307 pKa = 10.57SITAGCSSGLIAYY320 pKa = 7.74APPAGALWLRR330 pKa = 11.84EE331 pKa = 4.02PPLPQLFTPMPKK343 pKa = 9.94HH344 pKa = 6.29NKK346 pKa = 9.49ALLNPGEE353 pKa = 4.3VKK355 pKa = 10.18TNSLLATYY363 pKa = 9.94RR364 pKa = 11.84VRR366 pKa = 11.84NLDD369 pKa = 3.43FFKK372 pKa = 11.18SLFFGASTSLAKK384 pKa = 10.21DD385 pKa = 2.93VHH387 pKa = 5.84VQHH390 pKa = 7.08KK391 pKa = 9.34FGKK394 pKa = 8.53YY395 pKa = 9.82CFFALEE401 pKa = 4.05KK402 pKa = 9.77MLTVSAADD410 pKa = 3.56SSPQVGLEE418 pKa = 3.63LNQRR422 pKa = 11.84VGVMFTFPRR431 pKa = 11.84NTDD434 pKa = 3.02TAEE437 pKa = 3.84ITEE440 pKa = 4.48VPSFITPLTTT450 pKa = 3.83
MM1 pKa = 7.16SVVRR5 pKa = 11.84KK6 pKa = 8.61RR7 pKa = 11.84TGGYY11 pKa = 5.71TTYY14 pKa = 11.3APNKK18 pKa = 9.1KK19 pKa = 8.59MRR21 pKa = 11.84SGRR24 pKa = 11.84DD25 pKa = 3.15ILVSAGLAASRR36 pKa = 11.84KK37 pKa = 8.04YY38 pKa = 10.69GPRR41 pKa = 11.84LVSMAAQKK49 pKa = 9.93LASMAYY55 pKa = 9.8AAAKK59 pKa = 9.89DD60 pKa = 3.77DD61 pKa = 5.53PITQFRR67 pKa = 11.84NAKK70 pKa = 9.47KK71 pKa = 8.16RR72 pKa = 11.84TKK74 pKa = 9.92GRR76 pKa = 11.84SYY78 pKa = 11.28SEE80 pKa = 4.1QKK82 pKa = 10.59GVAAGKK88 pKa = 9.75FRR90 pKa = 11.84ATKK93 pKa = 10.36RR94 pKa = 11.84SPKK97 pKa = 10.35NIWLKK102 pKa = 7.72YY103 pKa = 6.6TRR105 pKa = 11.84KK106 pKa = 10.31GCVTTKK112 pKa = 10.37EE113 pKa = 4.29HH114 pKa = 6.32GCVLTGTVNTNAIYY128 pKa = 10.18FGHH131 pKa = 5.78STHH134 pKa = 7.04ANTYY138 pKa = 10.89LMLTVYY144 pKa = 9.48MRR146 pKa = 11.84AIVIHH151 pKa = 6.6IFRR154 pKa = 11.84KK155 pKa = 10.23GGVQIEE161 pKa = 4.65DD162 pKa = 4.12IEE164 pKa = 4.61TEE166 pKa = 4.31SPGSGGSMTIEE177 pKa = 3.73YY178 pKa = 8.73TYY180 pKa = 10.4NTDD183 pKa = 2.76IANQFVTVAFTGGLTLNQIAGSFASQFTNILFNNANDD220 pKa = 3.68EE221 pKa = 4.18QRR223 pKa = 11.84FFPILEE229 pKa = 4.27AKK231 pKa = 10.21IEE233 pKa = 4.06IPNLEE238 pKa = 4.56LCKK241 pKa = 9.93QDD243 pKa = 3.32YY244 pKa = 10.26RR245 pKa = 11.84NSIVHH250 pKa = 5.94VMVKK254 pKa = 10.38SDD256 pKa = 4.09LKK258 pKa = 9.95VQNRR262 pKa = 11.84TLATGTDD269 pKa = 3.45EE270 pKa = 5.82DD271 pKa = 4.59NNSSEE276 pKa = 4.85NIANQPLYY284 pKa = 11.16GKK286 pKa = 10.05FYY288 pKa = 9.59HH289 pKa = 6.78GKK291 pKa = 9.65GNGVFARR298 pKa = 11.84ANAGAGTGKK307 pKa = 10.57SITAGCSSGLIAYY320 pKa = 7.74APPAGALWLRR330 pKa = 11.84EE331 pKa = 4.02PPLPQLFTPMPKK343 pKa = 9.94HH344 pKa = 6.29NKK346 pKa = 9.49ALLNPGEE353 pKa = 4.3VKK355 pKa = 10.18TNSLLATYY363 pKa = 9.94RR364 pKa = 11.84VRR366 pKa = 11.84NLDD369 pKa = 3.43FFKK372 pKa = 11.18SLFFGASTSLAKK384 pKa = 10.21DD385 pKa = 2.93VHH387 pKa = 5.84VQHH390 pKa = 7.08KK391 pKa = 9.34FGKK394 pKa = 8.53YY395 pKa = 9.82CFFALEE401 pKa = 4.05KK402 pKa = 9.77MLTVSAADD410 pKa = 3.56SSPQVGLEE418 pKa = 3.63LNQRR422 pKa = 11.84VGVMFTFPRR431 pKa = 11.84NTDD434 pKa = 3.02TAEE437 pKa = 3.84ITEE440 pKa = 4.48VPSFITPLTTT450 pKa = 3.83
Molecular weight: 49.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
716 |
266 |
450 |
358.0 |
39.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.799 ± 0.757 | 1.117 ± 0.006 |
4.609 ± 1.498 | 4.33 ± 0.33 |
4.888 ± 0.445 | 7.961 ± 0.039 |
2.514 ± 0.514 | 5.447 ± 0.558 |
6.006 ± 1.106 | 7.542 ± 0.014 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.35 | 5.168 ± 1.055 |
4.609 ± 0.165 | 2.793 ± 0.54 |
6.983 ± 1.872 | 5.866 ± 0.801 |
7.821 ± 1.068 | 6.425 ± 0.425 |
1.536 ± 1.092 | 3.492 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
