
Sewage-associated circular DNA virus-34
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.03
Get precalculated fractions of proteins
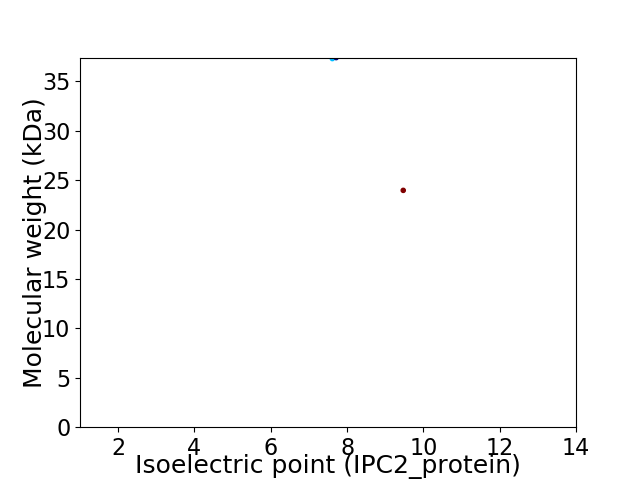
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH99|A0A0B4UH99_9VIRU Coat protein OS=Sewage-associated circular DNA virus-34 OX=1592101 PE=4 SV=1
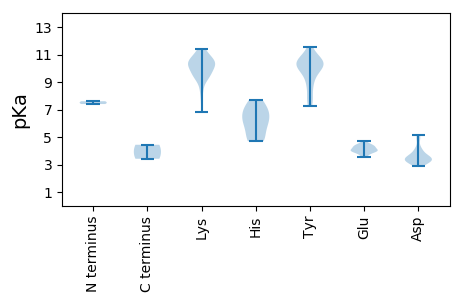
MM1 pKa = 7.6SKK3 pKa = 10.27PLRR6 pKa = 11.84NICFTSFNLDD16 pKa = 3.29IEE18 pKa = 4.46WFEE21 pKa = 4.49TEE23 pKa = 4.34WPSHH27 pKa = 5.76HH28 pKa = 7.01LPGEE32 pKa = 4.15EE33 pKa = 3.73TGMVQYY39 pKa = 10.57IIVQGEE45 pKa = 3.96FCKK48 pKa = 10.45EE49 pKa = 3.97GKK51 pKa = 9.69KK52 pKa = 10.05HH53 pKa = 4.84IQGYY57 pKa = 8.68AQLRR61 pKa = 11.84GQYY64 pKa = 8.14YY65 pKa = 10.08LKK67 pKa = 10.41KK68 pKa = 9.61IKK70 pKa = 10.43EE71 pKa = 4.1FFGDD75 pKa = 3.18NGLHH79 pKa = 5.03VEE81 pKa = 4.32PRR83 pKa = 11.84RR84 pKa = 11.84GTHH87 pKa = 4.7EE88 pKa = 3.87QARR91 pKa = 11.84DD92 pKa = 3.39YY93 pKa = 11.21CKK95 pKa = 10.16QEE97 pKa = 4.06KK98 pKa = 9.16NGRR101 pKa = 11.84WHH103 pKa = 7.7DD104 pKa = 3.93YY105 pKa = 11.54VEE107 pKa = 4.68MGDD110 pKa = 3.53QKK112 pKa = 11.42KK113 pKa = 10.17PGSRR117 pKa = 11.84TDD119 pKa = 3.61LLDD122 pKa = 4.13VIDD125 pKa = 5.01KK126 pKa = 10.37IKK128 pKa = 10.71EE129 pKa = 4.1GEE131 pKa = 4.31TIASIISNSRR141 pKa = 11.84DD142 pKa = 2.97PKK144 pKa = 8.44MVRR147 pKa = 11.84NAVVYY152 pKa = 10.66NRR154 pKa = 11.84TLTAIEE160 pKa = 4.25KK161 pKa = 9.29QFKK164 pKa = 9.44YY165 pKa = 10.69DD166 pKa = 3.49AAIKK170 pKa = 9.72EE171 pKa = 4.34AIKK174 pKa = 10.48EE175 pKa = 4.0YY176 pKa = 11.2DD177 pKa = 3.23GVTWRR182 pKa = 11.84KK183 pKa = 6.78WQEE186 pKa = 3.58EE187 pKa = 4.29LLCYY191 pKa = 9.92VDD193 pKa = 5.14KK194 pKa = 11.4DD195 pKa = 3.25ADD197 pKa = 3.53KK198 pKa = 11.25RR199 pKa = 11.84KK200 pKa = 9.42VRR202 pKa = 11.84WYY204 pKa = 9.73TDD206 pKa = 2.92MGGNTGKK213 pKa = 10.67SYY215 pKa = 9.13ITTYY219 pKa = 9.8LTLLGKK225 pKa = 10.46AYY227 pKa = 10.19VVSGGKK233 pKa = 9.07QADD236 pKa = 3.19ILYY239 pKa = 9.96AYY241 pKa = 8.71EE242 pKa = 4.38DD243 pKa = 4.04QPIVIYY249 pKa = 10.69DD250 pKa = 3.83LARR253 pKa = 11.84AYY255 pKa = 7.78EE256 pKa = 3.91QNMDD260 pKa = 4.17HH261 pKa = 7.06IYY263 pKa = 9.66VTMEE267 pKa = 3.53YY268 pKa = 10.17FKK270 pKa = 11.07NGRR273 pKa = 11.84FLSTKK278 pKa = 9.41YY279 pKa = 9.05EE280 pKa = 3.94SKK282 pKa = 9.73MRR284 pKa = 11.84VFKK287 pKa = 10.52RR288 pKa = 11.84PHH290 pKa = 6.2VIVMANFPPHH300 pKa = 6.3INKK303 pKa = 9.92LSEE306 pKa = 4.04DD307 pKa = 3.39RR308 pKa = 11.84WDD310 pKa = 3.27IVYY313 pKa = 7.48MTNPP317 pKa = 3.42
MM1 pKa = 7.6SKK3 pKa = 10.27PLRR6 pKa = 11.84NICFTSFNLDD16 pKa = 3.29IEE18 pKa = 4.46WFEE21 pKa = 4.49TEE23 pKa = 4.34WPSHH27 pKa = 5.76HH28 pKa = 7.01LPGEE32 pKa = 4.15EE33 pKa = 3.73TGMVQYY39 pKa = 10.57IIVQGEE45 pKa = 3.96FCKK48 pKa = 10.45EE49 pKa = 3.97GKK51 pKa = 9.69KK52 pKa = 10.05HH53 pKa = 4.84IQGYY57 pKa = 8.68AQLRR61 pKa = 11.84GQYY64 pKa = 8.14YY65 pKa = 10.08LKK67 pKa = 10.41KK68 pKa = 9.61IKK70 pKa = 10.43EE71 pKa = 4.1FFGDD75 pKa = 3.18NGLHH79 pKa = 5.03VEE81 pKa = 4.32PRR83 pKa = 11.84RR84 pKa = 11.84GTHH87 pKa = 4.7EE88 pKa = 3.87QARR91 pKa = 11.84DD92 pKa = 3.39YY93 pKa = 11.21CKK95 pKa = 10.16QEE97 pKa = 4.06KK98 pKa = 9.16NGRR101 pKa = 11.84WHH103 pKa = 7.7DD104 pKa = 3.93YY105 pKa = 11.54VEE107 pKa = 4.68MGDD110 pKa = 3.53QKK112 pKa = 11.42KK113 pKa = 10.17PGSRR117 pKa = 11.84TDD119 pKa = 3.61LLDD122 pKa = 4.13VIDD125 pKa = 5.01KK126 pKa = 10.37IKK128 pKa = 10.71EE129 pKa = 4.1GEE131 pKa = 4.31TIASIISNSRR141 pKa = 11.84DD142 pKa = 2.97PKK144 pKa = 8.44MVRR147 pKa = 11.84NAVVYY152 pKa = 10.66NRR154 pKa = 11.84TLTAIEE160 pKa = 4.25KK161 pKa = 9.29QFKK164 pKa = 9.44YY165 pKa = 10.69DD166 pKa = 3.49AAIKK170 pKa = 9.72EE171 pKa = 4.34AIKK174 pKa = 10.48EE175 pKa = 4.0YY176 pKa = 11.2DD177 pKa = 3.23GVTWRR182 pKa = 11.84KK183 pKa = 6.78WQEE186 pKa = 3.58EE187 pKa = 4.29LLCYY191 pKa = 9.92VDD193 pKa = 5.14KK194 pKa = 11.4DD195 pKa = 3.25ADD197 pKa = 3.53KK198 pKa = 11.25RR199 pKa = 11.84KK200 pKa = 9.42VRR202 pKa = 11.84WYY204 pKa = 9.73TDD206 pKa = 2.92MGGNTGKK213 pKa = 10.67SYY215 pKa = 9.13ITTYY219 pKa = 9.8LTLLGKK225 pKa = 10.46AYY227 pKa = 10.19VVSGGKK233 pKa = 9.07QADD236 pKa = 3.19ILYY239 pKa = 9.96AYY241 pKa = 8.71EE242 pKa = 4.38DD243 pKa = 4.04QPIVIYY249 pKa = 10.69DD250 pKa = 3.83LARR253 pKa = 11.84AYY255 pKa = 7.78EE256 pKa = 3.91QNMDD260 pKa = 4.17HH261 pKa = 7.06IYY263 pKa = 9.66VTMEE267 pKa = 3.53YY268 pKa = 10.17FKK270 pKa = 11.07NGRR273 pKa = 11.84FLSTKK278 pKa = 9.41YY279 pKa = 9.05EE280 pKa = 3.94SKK282 pKa = 9.73MRR284 pKa = 11.84VFKK287 pKa = 10.52RR288 pKa = 11.84PHH290 pKa = 6.2VIVMANFPPHH300 pKa = 6.3INKK303 pKa = 9.92LSEE306 pKa = 4.04DD307 pKa = 3.39RR308 pKa = 11.84WDD310 pKa = 3.27IVYY313 pKa = 7.48MTNPP317 pKa = 3.42
Molecular weight: 37.32 kDa
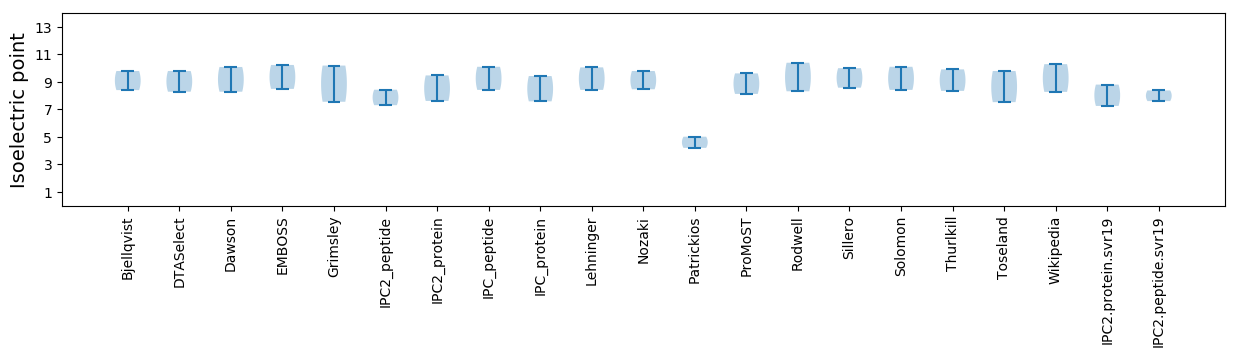
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH99|A0A0B4UH99_9VIRU Coat protein OS=Sewage-associated circular DNA virus-34 OX=1592101 PE=4 SV=1
MM1 pKa = 7.4SLVKK5 pKa = 10.65YY6 pKa = 10.27RR7 pKa = 11.84ISKK10 pKa = 9.68PKK12 pKa = 7.77YY13 pKa = 7.23TKK15 pKa = 10.28KK16 pKa = 10.42KK17 pKa = 9.21SEE19 pKa = 4.02KK20 pKa = 9.94KK21 pKa = 8.46FTSKK25 pKa = 10.69RR26 pKa = 11.84SINTMLSRR34 pKa = 11.84LPEE37 pKa = 3.79TKK39 pKa = 10.61YY40 pKa = 10.43YY41 pKa = 10.8DD42 pKa = 3.59VYY44 pKa = 10.42HH45 pKa = 6.13TTTITQNGALDD56 pKa = 4.79LLTGMTTLGTGEE68 pKa = 4.14GQRR71 pKa = 11.84VGSKK75 pKa = 10.23VFVKK79 pKa = 10.0YY80 pKa = 10.33IQVNGLIMRR89 pKa = 11.84NASSTIDD96 pKa = 3.36NLRR99 pKa = 11.84IIIFMDD105 pKa = 3.31KK106 pKa = 10.83QGMNTPAVTDD116 pKa = 3.41IMEE119 pKa = 4.49PAYY122 pKa = 10.38LGSGFSPVGRR132 pKa = 11.84INEE135 pKa = 3.95YY136 pKa = 10.31RR137 pKa = 11.84IPRR140 pKa = 11.84YY141 pKa = 9.89KK142 pKa = 9.99ILYY145 pKa = 9.29DD146 pKa = 3.46KK147 pKa = 9.46LTHH150 pKa = 6.98VSTQKK155 pKa = 10.96DD156 pKa = 3.56GFNFKK161 pKa = 11.16ANLRR165 pKa = 11.84INMPVYY171 pKa = 10.4YY172 pKa = 10.54VSTSTFKK179 pKa = 10.44NQVYY183 pKa = 10.47ILFLGDD189 pKa = 3.37NGNVLTLPYY198 pKa = 10.12CNYY201 pKa = 7.71TARR204 pKa = 11.84VSYY207 pKa = 10.52TDD209 pKa = 3.08QQ210 pKa = 4.44
MM1 pKa = 7.4SLVKK5 pKa = 10.65YY6 pKa = 10.27RR7 pKa = 11.84ISKK10 pKa = 9.68PKK12 pKa = 7.77YY13 pKa = 7.23TKK15 pKa = 10.28KK16 pKa = 10.42KK17 pKa = 9.21SEE19 pKa = 4.02KK20 pKa = 9.94KK21 pKa = 8.46FTSKK25 pKa = 10.69RR26 pKa = 11.84SINTMLSRR34 pKa = 11.84LPEE37 pKa = 3.79TKK39 pKa = 10.61YY40 pKa = 10.43YY41 pKa = 10.8DD42 pKa = 3.59VYY44 pKa = 10.42HH45 pKa = 6.13TTTITQNGALDD56 pKa = 4.79LLTGMTTLGTGEE68 pKa = 4.14GQRR71 pKa = 11.84VGSKK75 pKa = 10.23VFVKK79 pKa = 10.0YY80 pKa = 10.33IQVNGLIMRR89 pKa = 11.84NASSTIDD96 pKa = 3.36NLRR99 pKa = 11.84IIIFMDD105 pKa = 3.31KK106 pKa = 10.83QGMNTPAVTDD116 pKa = 3.41IMEE119 pKa = 4.49PAYY122 pKa = 10.38LGSGFSPVGRR132 pKa = 11.84INEE135 pKa = 3.95YY136 pKa = 10.31RR137 pKa = 11.84IPRR140 pKa = 11.84YY141 pKa = 9.89KK142 pKa = 9.99ILYY145 pKa = 9.29DD146 pKa = 3.46KK147 pKa = 9.46LTHH150 pKa = 6.98VSTQKK155 pKa = 10.96DD156 pKa = 3.56GFNFKK161 pKa = 11.16ANLRR165 pKa = 11.84INMPVYY171 pKa = 10.4YY172 pKa = 10.54VSTSTFKK179 pKa = 10.44NQVYY183 pKa = 10.47ILFLGDD189 pKa = 3.37NGNVLTLPYY198 pKa = 10.12CNYY201 pKa = 7.71TARR204 pKa = 11.84VSYY207 pKa = 10.52TDD209 pKa = 3.08QQ210 pKa = 4.44
Molecular weight: 23.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
527 |
210 |
317 |
263.5 |
30.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.985 ± 0.622 | 0.949 ± 0.26 |
5.693 ± 0.775 | 5.503 ± 1.721 |
3.605 ± 0.113 | 6.641 ± 0.014 |
2.087 ± 0.626 | 7.021 ± 0.067 |
9.108 ± 0.296 | 6.641 ± 0.801 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.416 ± 0.217 | 5.123 ± 0.851 |
3.605 ± 0.113 | 3.795 ± 0.254 |
5.503 ± 0.146 | 5.123 ± 1.113 |
7.4 ± 1.695 | 6.262 ± 0.223 |
1.328 ± 0.732 | 7.211 ± 0.225 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
