
Human papillomavirus type 128
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 13
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
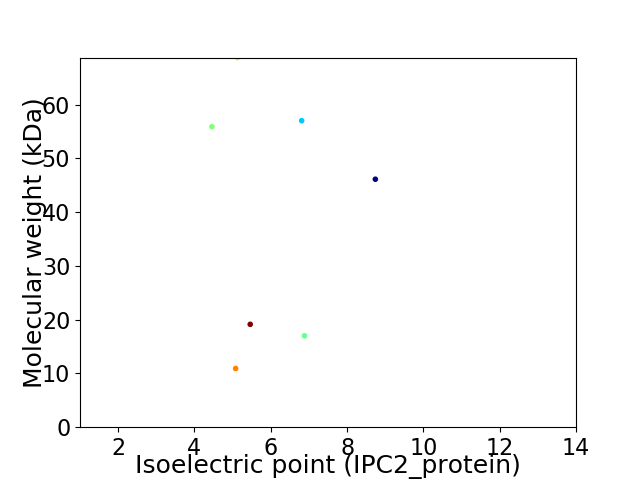
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7BRR3|E7BRR3_9PAPI Major capsid protein L1 OS=Human papillomavirus type 128 OX=931209 GN=L1 PE=3 SV=1
MM1 pKa = 7.47SKK3 pKa = 10.13PRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.6RR8 pKa = 11.84ASPEE12 pKa = 3.7DD13 pKa = 4.06LYY15 pKa = 11.46KK16 pKa = 10.8SCALGGDD23 pKa = 4.58CFPDD27 pKa = 3.44VQNKK31 pKa = 8.71IQGTTLADD39 pKa = 3.66RR40 pKa = 11.84LQVFGSILYY49 pKa = 10.33FGGLGIGSGKK59 pKa = 8.42GTGGATGYY67 pKa = 10.57RR68 pKa = 11.84PFGGGGTVSRR78 pKa = 11.84PSIRR82 pKa = 11.84IQPEE86 pKa = 4.21SLPKK90 pKa = 10.08PSVPIDD96 pKa = 3.59PLGGAEE102 pKa = 5.65LIPLDD107 pKa = 4.78VIDD110 pKa = 4.04ATAPSVVPLTEE121 pKa = 4.48GVPEE125 pKa = 4.04TTIISSGTGPTITAEE140 pKa = 4.13EE141 pKa = 4.65LDD143 pKa = 3.91VTTQIDD149 pKa = 4.63LNTFNNINEE158 pKa = 4.26HH159 pKa = 5.94PAVINVTDD167 pKa = 3.91EE168 pKa = 4.0GTTALDD174 pKa = 3.9VQTLPPPSKK183 pKa = 10.7KK184 pKa = 10.57LILDD188 pKa = 3.54STITDD193 pKa = 3.65QIEE196 pKa = 4.31LVPLHH201 pKa = 7.18RR202 pKa = 11.84IYY204 pKa = 10.88TDD206 pKa = 3.0QNVNMFVDD214 pKa = 3.58THH216 pKa = 6.72FEE218 pKa = 3.87GHH220 pKa = 6.12IVGGDD225 pKa = 3.44PNFTDD230 pKa = 2.86IVLRR234 pKa = 11.84EE235 pKa = 3.79QFEE238 pKa = 4.28IEE240 pKa = 4.21EE241 pKa = 4.28VSPKK245 pKa = 10.32TSTPTEE251 pKa = 4.11KK252 pKa = 10.78LSTAVGKK259 pKa = 10.31ARR261 pKa = 11.84RR262 pKa = 11.84VYY264 pKa = 10.76NRR266 pKa = 11.84FVKK269 pKa = 9.98QVPTSSLEE277 pKa = 3.97AVVNPSRR284 pKa = 11.84QVIFEE289 pKa = 4.13IEE291 pKa = 3.91NPAFDD296 pKa = 5.5DD297 pKa = 4.18EE298 pKa = 5.01VSLLFNQEE306 pKa = 3.88LAEE309 pKa = 4.36VTATPNPDD317 pKa = 3.09FRR319 pKa = 11.84DD320 pKa = 3.2INYY323 pKa = 9.78LSRR326 pKa = 11.84PFYY329 pKa = 11.11SEE331 pKa = 4.17TEE333 pKa = 4.13SGRR336 pKa = 11.84VQYY339 pKa = 11.13SRR341 pKa = 11.84LGQRR345 pKa = 11.84ASMQTRR351 pKa = 11.84SGLVIGEE358 pKa = 4.21KK359 pKa = 10.66VQFTYY364 pKa = 10.76NISDD368 pKa = 3.74IEE370 pKa = 4.03PVEE373 pKa = 4.23TIEE376 pKa = 5.66LSTLGTTSADD386 pKa = 3.77SIIQNAQAEE395 pKa = 4.91SVFIDD400 pKa = 4.05AQDD403 pKa = 3.9STPLTYY409 pKa = 10.53TEE411 pKa = 4.94DD412 pKa = 3.61QLLDD416 pKa = 3.66EE417 pKa = 5.28PNEE420 pKa = 4.31DD421 pKa = 3.27FANAHH426 pKa = 6.24LVLTTTDD433 pKa = 4.35DD434 pKa = 4.09FGDD437 pKa = 3.73TYY439 pKa = 10.64DD440 pKa = 4.26YY441 pKa = 11.27PVIPPGLGYY450 pKa = 11.02KK451 pKa = 10.4LLIPNYY457 pKa = 9.69NVTLSSFQLPEE468 pKa = 3.98ASVPTILPEE477 pKa = 4.3IPFAPYY483 pKa = 9.87APSPITVFGDD493 pKa = 3.59TFYY496 pKa = 11.49LNPSLLKK503 pKa = 10.31RR504 pKa = 11.84KK505 pKa = 9.51RR506 pKa = 11.84KK507 pKa = 9.25QYY509 pKa = 10.69FYY511 pKa = 11.61
MM1 pKa = 7.47SKK3 pKa = 10.13PRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.6RR8 pKa = 11.84ASPEE12 pKa = 3.7DD13 pKa = 4.06LYY15 pKa = 11.46KK16 pKa = 10.8SCALGGDD23 pKa = 4.58CFPDD27 pKa = 3.44VQNKK31 pKa = 8.71IQGTTLADD39 pKa = 3.66RR40 pKa = 11.84LQVFGSILYY49 pKa = 10.33FGGLGIGSGKK59 pKa = 8.42GTGGATGYY67 pKa = 10.57RR68 pKa = 11.84PFGGGGTVSRR78 pKa = 11.84PSIRR82 pKa = 11.84IQPEE86 pKa = 4.21SLPKK90 pKa = 10.08PSVPIDD96 pKa = 3.59PLGGAEE102 pKa = 5.65LIPLDD107 pKa = 4.78VIDD110 pKa = 4.04ATAPSVVPLTEE121 pKa = 4.48GVPEE125 pKa = 4.04TTIISSGTGPTITAEE140 pKa = 4.13EE141 pKa = 4.65LDD143 pKa = 3.91VTTQIDD149 pKa = 4.63LNTFNNINEE158 pKa = 4.26HH159 pKa = 5.94PAVINVTDD167 pKa = 3.91EE168 pKa = 4.0GTTALDD174 pKa = 3.9VQTLPPPSKK183 pKa = 10.7KK184 pKa = 10.57LILDD188 pKa = 3.54STITDD193 pKa = 3.65QIEE196 pKa = 4.31LVPLHH201 pKa = 7.18RR202 pKa = 11.84IYY204 pKa = 10.88TDD206 pKa = 3.0QNVNMFVDD214 pKa = 3.58THH216 pKa = 6.72FEE218 pKa = 3.87GHH220 pKa = 6.12IVGGDD225 pKa = 3.44PNFTDD230 pKa = 2.86IVLRR234 pKa = 11.84EE235 pKa = 3.79QFEE238 pKa = 4.28IEE240 pKa = 4.21EE241 pKa = 4.28VSPKK245 pKa = 10.32TSTPTEE251 pKa = 4.11KK252 pKa = 10.78LSTAVGKK259 pKa = 10.31ARR261 pKa = 11.84RR262 pKa = 11.84VYY264 pKa = 10.76NRR266 pKa = 11.84FVKK269 pKa = 9.98QVPTSSLEE277 pKa = 3.97AVVNPSRR284 pKa = 11.84QVIFEE289 pKa = 4.13IEE291 pKa = 3.91NPAFDD296 pKa = 5.5DD297 pKa = 4.18EE298 pKa = 5.01VSLLFNQEE306 pKa = 3.88LAEE309 pKa = 4.36VTATPNPDD317 pKa = 3.09FRR319 pKa = 11.84DD320 pKa = 3.2INYY323 pKa = 9.78LSRR326 pKa = 11.84PFYY329 pKa = 11.11SEE331 pKa = 4.17TEE333 pKa = 4.13SGRR336 pKa = 11.84VQYY339 pKa = 11.13SRR341 pKa = 11.84LGQRR345 pKa = 11.84ASMQTRR351 pKa = 11.84SGLVIGEE358 pKa = 4.21KK359 pKa = 10.66VQFTYY364 pKa = 10.76NISDD368 pKa = 3.74IEE370 pKa = 4.03PVEE373 pKa = 4.23TIEE376 pKa = 5.66LSTLGTTSADD386 pKa = 3.77SIIQNAQAEE395 pKa = 4.91SVFIDD400 pKa = 4.05AQDD403 pKa = 3.9STPLTYY409 pKa = 10.53TEE411 pKa = 4.94DD412 pKa = 3.61QLLDD416 pKa = 3.66EE417 pKa = 5.28PNEE420 pKa = 4.31DD421 pKa = 3.27FANAHH426 pKa = 6.24LVLTTTDD433 pKa = 4.35DD434 pKa = 4.09FGDD437 pKa = 3.73TYY439 pKa = 10.64DD440 pKa = 4.26YY441 pKa = 11.27PVIPPGLGYY450 pKa = 11.02KK451 pKa = 10.4LLIPNYY457 pKa = 9.69NVTLSSFQLPEE468 pKa = 3.98ASVPTILPEE477 pKa = 4.3IPFAPYY483 pKa = 9.87APSPITVFGDD493 pKa = 3.59TFYY496 pKa = 11.49LNPSLLKK503 pKa = 10.31RR504 pKa = 11.84KK505 pKa = 9.51RR506 pKa = 11.84KK507 pKa = 9.25QYY509 pKa = 10.69FYY511 pKa = 11.61
Molecular weight: 55.91 kDa
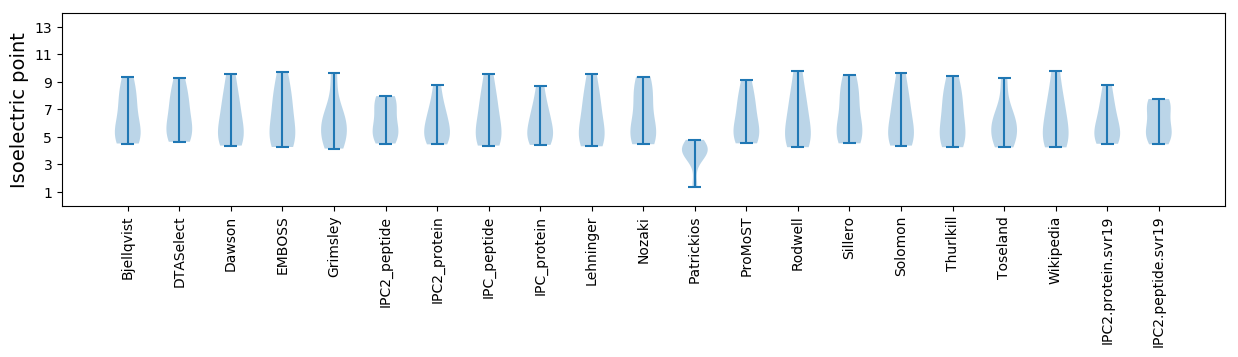
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7BRR1|E7BRR1_9PAPI E4 OS=Human papillomavirus type 128 OX=931209 GN=E4 PE=4 SV=1
MM1 pKa = 6.98NQAEE5 pKa = 4.38LTRR8 pKa = 11.84RR9 pKa = 11.84FDD11 pKa = 3.79ALQDD15 pKa = 3.67EE16 pKa = 4.46QLNLYY21 pKa = 7.5EE22 pKa = 4.64TGADD26 pKa = 3.75DD27 pKa = 4.0LQSQIRR33 pKa = 11.84HH34 pKa = 5.39WEE36 pKa = 3.74LTKK39 pKa = 10.68QLNLYY44 pKa = 8.16MFYY47 pKa = 10.83GRR49 pKa = 11.84KK50 pKa = 9.24EE51 pKa = 4.14GYY53 pKa = 8.93TNFGMQRR60 pKa = 11.84LPILQVSEE68 pKa = 4.49YY69 pKa = 9.33NAKK72 pKa = 9.98IAINMLILLKK82 pKa = 10.81SLANSKK88 pKa = 10.67FKK90 pKa = 10.81DD91 pKa = 3.96EE92 pKa = 4.61KK93 pKa = 9.25WTLSDD98 pKa = 3.42TSSDD102 pKa = 4.27LMLTPPRR109 pKa = 11.84NTFKK113 pKa = 10.94KK114 pKa = 9.62SAYY117 pKa = 6.62QVKK120 pKa = 9.67VEE122 pKa = 4.47FDD124 pKa = 3.42NDD126 pKa = 3.65PEE128 pKa = 4.52NFFLYY133 pKa = 10.14TNWDD137 pKa = 3.58EE138 pKa = 5.89IYY140 pKa = 11.11YY141 pKa = 10.26QDD143 pKa = 6.23LSDD146 pKa = 4.39KK147 pKa = 8.33WHH149 pKa = 5.89KK150 pKa = 8.56TQGKK154 pKa = 8.73VDD156 pKa = 3.65HH157 pKa = 6.93NGLYY161 pKa = 10.43FDD163 pKa = 6.32DD164 pKa = 4.37ISGDD168 pKa = 3.26RR169 pKa = 11.84NYY171 pKa = 10.86FLLFSPEE178 pKa = 3.81SEE180 pKa = 4.44KK181 pKa = 11.19YY182 pKa = 10.7GKK184 pKa = 8.24TGLWTVHH191 pKa = 5.75YY192 pKa = 9.76KK193 pKa = 9.06STTISSVVTSSSKK206 pKa = 10.61LSSGGYY212 pKa = 9.29SGQPSDD218 pKa = 3.53ISRR221 pKa = 11.84SSSRR225 pKa = 11.84CSSPQEE231 pKa = 3.96GTSRR235 pKa = 11.84RR236 pKa = 11.84CEE238 pKa = 3.73EE239 pKa = 3.92TRR241 pKa = 11.84QKK243 pKa = 11.03GSSYY247 pKa = 11.04DD248 pKa = 3.66SPTSTVGSGRR258 pKa = 11.84GKK260 pKa = 10.17RR261 pKa = 11.84KK262 pKa = 9.89RR263 pKa = 11.84EE264 pKa = 4.11SEE266 pKa = 4.22SKK268 pKa = 10.51SPSRR272 pKa = 11.84RR273 pKa = 11.84PARR276 pKa = 11.84KK277 pKa = 9.0RR278 pKa = 11.84RR279 pKa = 11.84VHH281 pKa = 6.66SEE283 pKa = 4.08LSPVSAEE290 pKa = 3.98EE291 pKa = 3.44VGRR294 pKa = 11.84RR295 pKa = 11.84HH296 pKa = 6.67RR297 pKa = 11.84SLPRR301 pKa = 11.84HH302 pKa = 5.13GLSRR306 pKa = 11.84LGRR309 pKa = 11.84LEE311 pKa = 4.52AEE313 pKa = 4.24ARR315 pKa = 11.84DD316 pKa = 3.91PDD318 pKa = 3.85VLLVKK323 pKa = 10.79GSANNLKK330 pKa = 9.67CWRR333 pKa = 11.84YY334 pKa = 9.0RR335 pKa = 11.84CEE337 pKa = 4.13KK338 pKa = 10.65KK339 pKa = 10.43FGHH342 pKa = 6.76LFSFISSVFRR352 pKa = 11.84WINDD356 pKa = 3.4DD357 pKa = 4.78FSQHH361 pKa = 5.2YY362 pKa = 10.08NSRR365 pKa = 11.84LLVAFEE371 pKa = 4.97NRR373 pKa = 11.84DD374 pKa = 3.34QRR376 pKa = 11.84NQFLMSVTFPKK387 pKa = 10.16QSSYY391 pKa = 11.05VFGSLDD397 pKa = 3.37SLL399 pKa = 4.17
MM1 pKa = 6.98NQAEE5 pKa = 4.38LTRR8 pKa = 11.84RR9 pKa = 11.84FDD11 pKa = 3.79ALQDD15 pKa = 3.67EE16 pKa = 4.46QLNLYY21 pKa = 7.5EE22 pKa = 4.64TGADD26 pKa = 3.75DD27 pKa = 4.0LQSQIRR33 pKa = 11.84HH34 pKa = 5.39WEE36 pKa = 3.74LTKK39 pKa = 10.68QLNLYY44 pKa = 8.16MFYY47 pKa = 10.83GRR49 pKa = 11.84KK50 pKa = 9.24EE51 pKa = 4.14GYY53 pKa = 8.93TNFGMQRR60 pKa = 11.84LPILQVSEE68 pKa = 4.49YY69 pKa = 9.33NAKK72 pKa = 9.98IAINMLILLKK82 pKa = 10.81SLANSKK88 pKa = 10.67FKK90 pKa = 10.81DD91 pKa = 3.96EE92 pKa = 4.61KK93 pKa = 9.25WTLSDD98 pKa = 3.42TSSDD102 pKa = 4.27LMLTPPRR109 pKa = 11.84NTFKK113 pKa = 10.94KK114 pKa = 9.62SAYY117 pKa = 6.62QVKK120 pKa = 9.67VEE122 pKa = 4.47FDD124 pKa = 3.42NDD126 pKa = 3.65PEE128 pKa = 4.52NFFLYY133 pKa = 10.14TNWDD137 pKa = 3.58EE138 pKa = 5.89IYY140 pKa = 11.11YY141 pKa = 10.26QDD143 pKa = 6.23LSDD146 pKa = 4.39KK147 pKa = 8.33WHH149 pKa = 5.89KK150 pKa = 8.56TQGKK154 pKa = 8.73VDD156 pKa = 3.65HH157 pKa = 6.93NGLYY161 pKa = 10.43FDD163 pKa = 6.32DD164 pKa = 4.37ISGDD168 pKa = 3.26RR169 pKa = 11.84NYY171 pKa = 10.86FLLFSPEE178 pKa = 3.81SEE180 pKa = 4.44KK181 pKa = 11.19YY182 pKa = 10.7GKK184 pKa = 8.24TGLWTVHH191 pKa = 5.75YY192 pKa = 9.76KK193 pKa = 9.06STTISSVVTSSSKK206 pKa = 10.61LSSGGYY212 pKa = 9.29SGQPSDD218 pKa = 3.53ISRR221 pKa = 11.84SSSRR225 pKa = 11.84CSSPQEE231 pKa = 3.96GTSRR235 pKa = 11.84RR236 pKa = 11.84CEE238 pKa = 3.73EE239 pKa = 3.92TRR241 pKa = 11.84QKK243 pKa = 11.03GSSYY247 pKa = 11.04DD248 pKa = 3.66SPTSTVGSGRR258 pKa = 11.84GKK260 pKa = 10.17RR261 pKa = 11.84KK262 pKa = 9.89RR263 pKa = 11.84EE264 pKa = 4.11SEE266 pKa = 4.22SKK268 pKa = 10.51SPSRR272 pKa = 11.84RR273 pKa = 11.84PARR276 pKa = 11.84KK277 pKa = 9.0RR278 pKa = 11.84RR279 pKa = 11.84VHH281 pKa = 6.66SEE283 pKa = 4.08LSPVSAEE290 pKa = 3.98EE291 pKa = 3.44VGRR294 pKa = 11.84RR295 pKa = 11.84HH296 pKa = 6.67RR297 pKa = 11.84SLPRR301 pKa = 11.84HH302 pKa = 5.13GLSRR306 pKa = 11.84LGRR309 pKa = 11.84LEE311 pKa = 4.52AEE313 pKa = 4.24ARR315 pKa = 11.84DD316 pKa = 3.91PDD318 pKa = 3.85VLLVKK323 pKa = 10.79GSANNLKK330 pKa = 9.67CWRR333 pKa = 11.84YY334 pKa = 9.0RR335 pKa = 11.84CEE337 pKa = 4.13KK338 pKa = 10.65KK339 pKa = 10.43FGHH342 pKa = 6.76LFSFISSVFRR352 pKa = 11.84WINDD356 pKa = 3.4DD357 pKa = 4.78FSQHH361 pKa = 5.2YY362 pKa = 10.08NSRR365 pKa = 11.84LLVAFEE371 pKa = 4.97NRR373 pKa = 11.84DD374 pKa = 3.34QRR376 pKa = 11.84NQFLMSVTFPKK387 pKa = 10.16QSSYY391 pKa = 11.05VFGSLDD397 pKa = 3.37SLL399 pKa = 4.17
Molecular weight: 46.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2421 |
96 |
605 |
345.9 |
39.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.915 ± 0.561 | 2.23 ± 0.763 |
6.526 ± 0.221 | 6.072 ± 0.345 |
5.081 ± 0.388 | 5.411 ± 0.528 |
1.817 ± 0.263 | 4.915 ± 0.61 |
5.535 ± 0.581 | 9.748 ± 0.62 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.652 ± 0.26 | 5.659 ± 0.588 |
5.535 ± 1.012 | 4.833 ± 0.327 |
5.411 ± 0.832 | 7.972 ± 1.171 |
6.237 ± 0.827 | 5.659 ± 0.612 |
1.239 ± 0.275 | 3.552 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
