
Porcine associated porprismacovirus 8
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
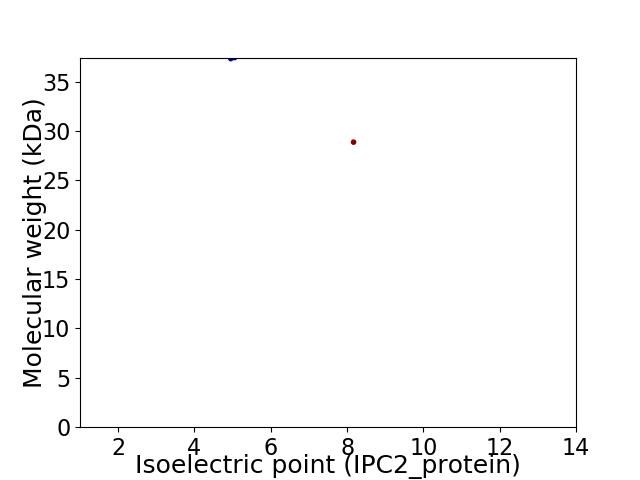
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
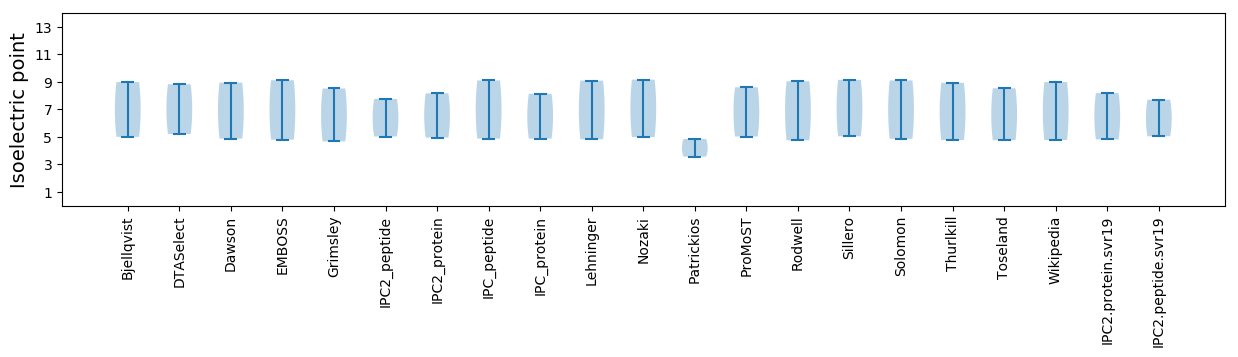
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076VH53|A0A076VH53_9VIRU Cap protein OS=Porcine associated porprismacovirus 8 OX=2170124 PE=4 SV=1
MM1 pKa = 8.17VMVRR5 pKa = 11.84VSEE8 pKa = 4.34TYY10 pKa = 10.7DD11 pKa = 4.06LSTKK15 pKa = 6.66TNKK18 pKa = 9.32MGIVGIHH25 pKa = 5.16TPRR28 pKa = 11.84GNLIDD33 pKa = 3.94QMWPGLVLQHH43 pKa = 6.23KK44 pKa = 9.87KK45 pKa = 10.34FRR47 pKa = 11.84FVQCDD52 pKa = 3.04VAMACASMLPADD64 pKa = 4.26PLQIGVEE71 pKa = 4.06AGAIAPQDD79 pKa = 3.54MFNPILYY86 pKa = 9.77KK87 pKa = 10.6AVSNDD92 pKa = 2.66SMSQVLQYY100 pKa = 10.65LAQAQQTGSGTTNTGLLQGSVASVNDD126 pKa = 3.34VAFKK130 pKa = 10.99ADD132 pKa = 4.13AEE134 pKa = 4.38DD135 pKa = 3.97TVNIDD140 pKa = 3.57QFNIYY145 pKa = 10.03YY146 pKa = 10.77GLLADD151 pKa = 3.65SDD153 pKa = 4.19GWRR156 pKa = 11.84KK157 pKa = 10.0AMPQAGLEE165 pKa = 4.04MRR167 pKa = 11.84GLYY170 pKa = 9.89PLVFSVMSQFGTNGYY185 pKa = 5.65TTGEE189 pKa = 4.34GVSGGTMDD197 pKa = 5.4AEE199 pKa = 4.59SLNHH203 pKa = 5.87QYY205 pKa = 11.33GMSASSDD212 pKa = 2.82LSTYY216 pKa = 10.52RR217 pKa = 11.84NSLMRR222 pKa = 11.84GPTMRR227 pKa = 11.84MPALDD232 pKa = 3.78TVCYY236 pKa = 10.2NPNDD240 pKa = 3.66VSSGDD245 pKa = 3.6VVPNSNALVHH255 pKa = 6.56SNVGEE260 pKa = 4.07VPPAYY265 pKa = 10.15VGLIVLPPAKK275 pKa = 10.23LNQLYY280 pKa = 10.26YY281 pKa = 10.12RR282 pKa = 11.84LKK284 pKa = 9.08VTWTVEE290 pKa = 4.01FSGLRR295 pKa = 11.84SMNDD299 pKa = 2.73IAKK302 pKa = 8.87WAMLGRR308 pKa = 11.84IGSSSYY314 pKa = 9.5GTDD317 pKa = 3.04YY318 pKa = 11.69ANQSRR323 pKa = 11.84SMPEE327 pKa = 3.25QTAMVDD333 pKa = 3.14TGGAAIEE340 pKa = 4.8KK341 pKa = 10.05IMEE344 pKa = 4.37GAA346 pKa = 3.51
MM1 pKa = 8.17VMVRR5 pKa = 11.84VSEE8 pKa = 4.34TYY10 pKa = 10.7DD11 pKa = 4.06LSTKK15 pKa = 6.66TNKK18 pKa = 9.32MGIVGIHH25 pKa = 5.16TPRR28 pKa = 11.84GNLIDD33 pKa = 3.94QMWPGLVLQHH43 pKa = 6.23KK44 pKa = 9.87KK45 pKa = 10.34FRR47 pKa = 11.84FVQCDD52 pKa = 3.04VAMACASMLPADD64 pKa = 4.26PLQIGVEE71 pKa = 4.06AGAIAPQDD79 pKa = 3.54MFNPILYY86 pKa = 9.77KK87 pKa = 10.6AVSNDD92 pKa = 2.66SMSQVLQYY100 pKa = 10.65LAQAQQTGSGTTNTGLLQGSVASVNDD126 pKa = 3.34VAFKK130 pKa = 10.99ADD132 pKa = 4.13AEE134 pKa = 4.38DD135 pKa = 3.97TVNIDD140 pKa = 3.57QFNIYY145 pKa = 10.03YY146 pKa = 10.77GLLADD151 pKa = 3.65SDD153 pKa = 4.19GWRR156 pKa = 11.84KK157 pKa = 10.0AMPQAGLEE165 pKa = 4.04MRR167 pKa = 11.84GLYY170 pKa = 9.89PLVFSVMSQFGTNGYY185 pKa = 5.65TTGEE189 pKa = 4.34GVSGGTMDD197 pKa = 5.4AEE199 pKa = 4.59SLNHH203 pKa = 5.87QYY205 pKa = 11.33GMSASSDD212 pKa = 2.82LSTYY216 pKa = 10.52RR217 pKa = 11.84NSLMRR222 pKa = 11.84GPTMRR227 pKa = 11.84MPALDD232 pKa = 3.78TVCYY236 pKa = 10.2NPNDD240 pKa = 3.66VSSGDD245 pKa = 3.6VVPNSNALVHH255 pKa = 6.56SNVGEE260 pKa = 4.07VPPAYY265 pKa = 10.15VGLIVLPPAKK275 pKa = 10.23LNQLYY280 pKa = 10.26YY281 pKa = 10.12RR282 pKa = 11.84LKK284 pKa = 9.08VTWTVEE290 pKa = 4.01FSGLRR295 pKa = 11.84SMNDD299 pKa = 2.73IAKK302 pKa = 8.87WAMLGRR308 pKa = 11.84IGSSSYY314 pKa = 9.5GTDD317 pKa = 3.04YY318 pKa = 11.69ANQSRR323 pKa = 11.84SMPEE327 pKa = 3.25QTAMVDD333 pKa = 3.14TGGAAIEE340 pKa = 4.8KK341 pKa = 10.05IMEE344 pKa = 4.37GAA346 pKa = 3.51
Molecular weight: 37.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076VH53|A0A076VH53_9VIRU Cap protein OS=Porcine associated porprismacovirus 8 OX=2170124 PE=4 SV=1
MM1 pKa = 6.91KK2 pKa = 9.8TYY4 pKa = 9.95MLTIPRR10 pKa = 11.84SVPKK14 pKa = 10.05RR15 pKa = 11.84AIMTMIEE22 pKa = 4.06VNDD25 pKa = 3.84CKK27 pKa = 10.59RR28 pKa = 11.84WIVAKK33 pKa = 8.98EE34 pKa = 4.14TGHH37 pKa = 6.63GGYY40 pKa = 10.08DD41 pKa = 2.71HH42 pKa = 6.47WQIRR46 pKa = 11.84LQTSNDD52 pKa = 3.92SFFEE56 pKa = 3.86WCEE59 pKa = 3.64QHH61 pKa = 6.95IPTAHH66 pKa = 6.48VEE68 pKa = 3.93EE69 pKa = 4.98AQNKK73 pKa = 7.47WEE75 pKa = 4.04YY76 pKa = 9.59EE77 pKa = 4.06RR78 pKa = 11.84KK79 pKa = 9.81EE80 pKa = 4.02GRR82 pKa = 11.84FWTSDD87 pKa = 3.19DD88 pKa = 3.43TTEE91 pKa = 3.99IRR93 pKa = 11.84IEE95 pKa = 4.0RR96 pKa = 11.84FGRR99 pKa = 11.84PRR101 pKa = 11.84GWQEE105 pKa = 3.45TALKK109 pKa = 10.41RR110 pKa = 11.84LRR112 pKa = 11.84TQNDD116 pKa = 3.26RR117 pKa = 11.84QVDD120 pKa = 3.36VWYY123 pKa = 10.2DD124 pKa = 3.53PIGNRR129 pKa = 11.84GKK131 pKa = 10.28SWLAGHH137 pKa = 7.38LFEE140 pKa = 5.26TGKK143 pKa = 10.86ACIVPRR149 pKa = 11.84DD150 pKa = 3.54WTNPSEE156 pKa = 3.9ICNYY160 pKa = 9.93LVHH163 pKa = 6.97NYY165 pKa = 9.94DD166 pKa = 3.58NEE168 pKa = 4.63PIIVIDD174 pKa = 3.74IPRR177 pKa = 11.84EE178 pKa = 3.68TGIPKK183 pKa = 10.24GFYY186 pKa = 9.63ATVEE190 pKa = 4.2MIKK193 pKa = 10.63DD194 pKa = 3.64GLIGTTKK201 pKa = 9.84WEE203 pKa = 3.77GRR205 pKa = 11.84MRR207 pKa = 11.84NIRR210 pKa = 11.84GVKK213 pKa = 9.23IIVMTNHH220 pKa = 6.04QMDD223 pKa = 4.66LKK225 pKa = 10.83KK226 pKa = 10.59LSKK229 pKa = 10.68DD230 pKa = 2.65RR231 pKa = 11.84WRR233 pKa = 11.84LNGINAQNAPMLL245 pKa = 4.27
MM1 pKa = 6.91KK2 pKa = 9.8TYY4 pKa = 9.95MLTIPRR10 pKa = 11.84SVPKK14 pKa = 10.05RR15 pKa = 11.84AIMTMIEE22 pKa = 4.06VNDD25 pKa = 3.84CKK27 pKa = 10.59RR28 pKa = 11.84WIVAKK33 pKa = 8.98EE34 pKa = 4.14TGHH37 pKa = 6.63GGYY40 pKa = 10.08DD41 pKa = 2.71HH42 pKa = 6.47WQIRR46 pKa = 11.84LQTSNDD52 pKa = 3.92SFFEE56 pKa = 3.86WCEE59 pKa = 3.64QHH61 pKa = 6.95IPTAHH66 pKa = 6.48VEE68 pKa = 3.93EE69 pKa = 4.98AQNKK73 pKa = 7.47WEE75 pKa = 4.04YY76 pKa = 9.59EE77 pKa = 4.06RR78 pKa = 11.84KK79 pKa = 9.81EE80 pKa = 4.02GRR82 pKa = 11.84FWTSDD87 pKa = 3.19DD88 pKa = 3.43TTEE91 pKa = 3.99IRR93 pKa = 11.84IEE95 pKa = 4.0RR96 pKa = 11.84FGRR99 pKa = 11.84PRR101 pKa = 11.84GWQEE105 pKa = 3.45TALKK109 pKa = 10.41RR110 pKa = 11.84LRR112 pKa = 11.84TQNDD116 pKa = 3.26RR117 pKa = 11.84QVDD120 pKa = 3.36VWYY123 pKa = 10.2DD124 pKa = 3.53PIGNRR129 pKa = 11.84GKK131 pKa = 10.28SWLAGHH137 pKa = 7.38LFEE140 pKa = 5.26TGKK143 pKa = 10.86ACIVPRR149 pKa = 11.84DD150 pKa = 3.54WTNPSEE156 pKa = 3.9ICNYY160 pKa = 9.93LVHH163 pKa = 6.97NYY165 pKa = 9.94DD166 pKa = 3.58NEE168 pKa = 4.63PIIVIDD174 pKa = 3.74IPRR177 pKa = 11.84EE178 pKa = 3.68TGIPKK183 pKa = 10.24GFYY186 pKa = 9.63ATVEE190 pKa = 4.2MIKK193 pKa = 10.63DD194 pKa = 3.64GLIGTTKK201 pKa = 9.84WEE203 pKa = 3.77GRR205 pKa = 11.84MRR207 pKa = 11.84NIRR210 pKa = 11.84GVKK213 pKa = 9.23IIVMTNHH220 pKa = 6.04QMDD223 pKa = 4.66LKK225 pKa = 10.83KK226 pKa = 10.59LSKK229 pKa = 10.68DD230 pKa = 2.65RR231 pKa = 11.84WRR233 pKa = 11.84LNGINAQNAPMLL245 pKa = 4.27
Molecular weight: 28.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
591 |
245 |
346 |
295.5 |
33.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.768 ± 1.66 | 1.184 ± 0.277 |
5.753 ± 0.024 | 4.907 ± 1.508 |
2.369 ± 0.05 | 8.291 ± 0.836 |
1.861 ± 0.615 | 5.922 ± 1.889 |
4.569 ± 1.213 | 6.768 ± 1.156 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
5.076 ± 0.867 | 5.584 ± 0.081 |
4.738 ± 0.153 | 4.569 ± 0.553 |
5.584 ± 1.846 | 6.261 ± 2.103 |
6.599 ± 0.462 | 6.937 ± 1.26 |
2.538 ± 1.206 | 3.723 ± 0.535 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
