
Emiliania huxleyi virus 86 (isolate United Kingdom/English Channel/1999) (EhV-86)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Algavirales; Phycodnaviridae; Coccolithovirus; Emiliania huxleyi virus 86
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
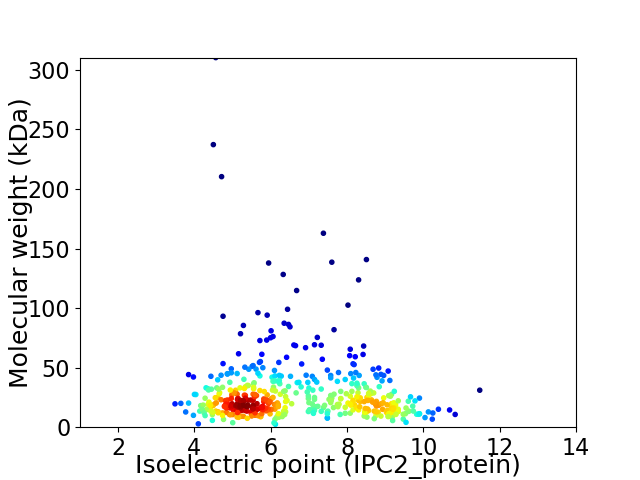
Virtual 2D-PAGE plot for 472 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
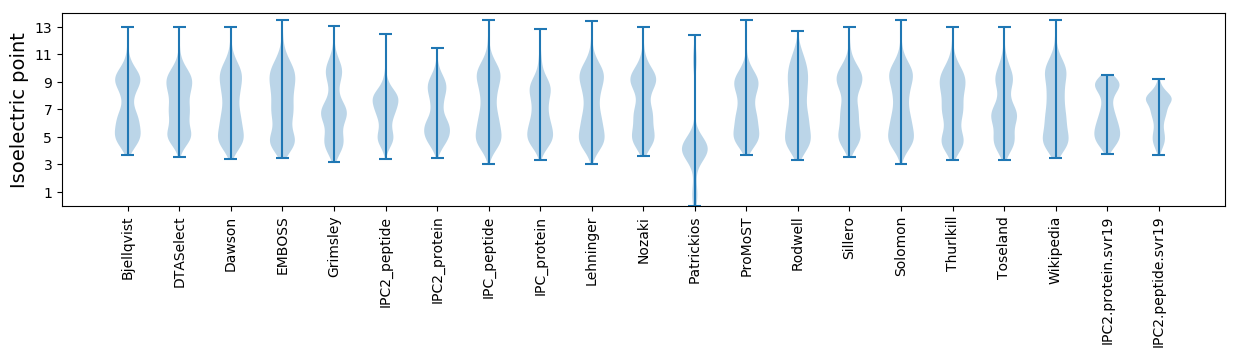
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4A372|Q4A372_EHV8U Sphingolipid 4-desaturase OS=Emiliania huxleyi virus 86 (isolate United Kingdom/English Channel/1999) OX=654925 GN=EhV061 PE=3 SV=1
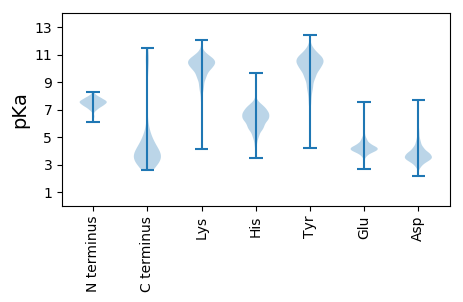
MM1 pKa = 7.24SVCKK5 pKa = 9.37ITNDD9 pKa = 5.27GIDD12 pKa = 3.35LANNVDD18 pKa = 3.48VDD20 pKa = 3.81TLYY23 pKa = 10.92KK24 pKa = 10.23RR25 pKa = 11.84EE26 pKa = 4.05EE27 pKa = 4.27VVIQPPASSLRR38 pKa = 11.84ICLYY42 pKa = 10.46TLDD45 pKa = 4.67KK46 pKa = 10.91PGTPVKK52 pKa = 10.56QSTLLSYY59 pKa = 10.74DD60 pKa = 3.94LPPPFDD66 pKa = 3.56NYY68 pKa = 11.01LYY70 pKa = 10.48NSPVYY75 pKa = 9.16MQFIGGTITSVSQATQLIKK94 pKa = 10.89LLGEE98 pKa = 4.36IFVHH102 pKa = 6.39TDD104 pKa = 3.19DD105 pKa = 5.45SVSDD109 pKa = 4.12SNTLVYY115 pKa = 10.62EE116 pKa = 4.0KK117 pKa = 11.17VEE119 pKa = 4.08DD120 pKa = 3.9VEE122 pKa = 4.77SEE124 pKa = 4.13EE125 pKa = 4.5EE126 pKa = 4.23EE127 pKa = 4.12EE128 pKa = 4.62DD129 pKa = 4.25DD130 pKa = 5.39VPEE133 pKa = 4.38EE134 pKa = 4.38YY135 pKa = 10.56YY136 pKa = 11.14DD137 pKa = 3.68HH138 pKa = 7.26NEE140 pKa = 3.67GSEE143 pKa = 3.99YY144 pKa = 11.02EE145 pKa = 4.05EE146 pKa = 5.84DD147 pKa = 3.54EE148 pKa = 4.33DD149 pKa = 4.59VAYY152 pKa = 10.95SDD154 pKa = 6.17DD155 pKa = 4.07EE156 pKa = 6.76DD157 pKa = 5.97DD158 pKa = 5.8DD159 pKa = 4.32EE160 pKa = 6.81SKK162 pKa = 11.31VEE164 pKa = 4.4DD165 pKa = 3.66NSTVYY170 pKa = 10.89EE171 pKa = 4.63DD172 pKa = 5.38DD173 pKa = 4.39SSLKK177 pKa = 10.73
MM1 pKa = 7.24SVCKK5 pKa = 9.37ITNDD9 pKa = 5.27GIDD12 pKa = 3.35LANNVDD18 pKa = 3.48VDD20 pKa = 3.81TLYY23 pKa = 10.92KK24 pKa = 10.23RR25 pKa = 11.84EE26 pKa = 4.05EE27 pKa = 4.27VVIQPPASSLRR38 pKa = 11.84ICLYY42 pKa = 10.46TLDD45 pKa = 4.67KK46 pKa = 10.91PGTPVKK52 pKa = 10.56QSTLLSYY59 pKa = 10.74DD60 pKa = 3.94LPPPFDD66 pKa = 3.56NYY68 pKa = 11.01LYY70 pKa = 10.48NSPVYY75 pKa = 9.16MQFIGGTITSVSQATQLIKK94 pKa = 10.89LLGEE98 pKa = 4.36IFVHH102 pKa = 6.39TDD104 pKa = 3.19DD105 pKa = 5.45SVSDD109 pKa = 4.12SNTLVYY115 pKa = 10.62EE116 pKa = 4.0KK117 pKa = 11.17VEE119 pKa = 4.08DD120 pKa = 3.9VEE122 pKa = 4.77SEE124 pKa = 4.13EE125 pKa = 4.5EE126 pKa = 4.23EE127 pKa = 4.12EE128 pKa = 4.62DD129 pKa = 4.25DD130 pKa = 5.39VPEE133 pKa = 4.38EE134 pKa = 4.38YY135 pKa = 10.56YY136 pKa = 11.14DD137 pKa = 3.68HH138 pKa = 7.26NEE140 pKa = 3.67GSEE143 pKa = 3.99YY144 pKa = 11.02EE145 pKa = 4.05EE146 pKa = 5.84DD147 pKa = 3.54EE148 pKa = 4.33DD149 pKa = 4.59VAYY152 pKa = 10.95SDD154 pKa = 6.17DD155 pKa = 4.07EE156 pKa = 6.76DD157 pKa = 5.97DD158 pKa = 5.8DD159 pKa = 4.32EE160 pKa = 6.81SKK162 pKa = 11.31VEE164 pKa = 4.4DD165 pKa = 3.66NSTVYY170 pKa = 10.89EE171 pKa = 4.63DD172 pKa = 5.38DD173 pKa = 4.39SSLKK177 pKa = 10.73
Molecular weight: 20.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4A2V5|Q4A2V5_EHV8U Major Facilitator Superfamily protein OS=Emiliania huxleyi virus 86 (isolate United Kingdom/English Channel/1999) OX=654925 GN=EhV179 PE=4 SV=1
MM1 pKa = 7.25AVSRR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84YY9 pKa = 7.36GTVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TVRR20 pKa = 11.84KK21 pKa = 8.12STASRR26 pKa = 11.84STKK29 pKa = 9.49VGRR32 pKa = 11.84VAKK35 pKa = 9.99LSNMSAGCRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.55VKK48 pKa = 10.54SQLGNRR54 pKa = 11.84KK55 pKa = 9.43LSRR58 pKa = 11.84ATRR61 pKa = 11.84ASIIRR66 pKa = 11.84RR67 pKa = 11.84SVKK70 pKa = 8.22TWSRR74 pKa = 11.84MSKK77 pKa = 9.88ARR79 pKa = 11.84KK80 pKa = 8.16SRR82 pKa = 11.84YY83 pKa = 5.01MTRR86 pKa = 11.84KK87 pKa = 8.7SSKK90 pKa = 10.01SKK92 pKa = 10.52RR93 pKa = 11.84KK94 pKa = 4.15TTRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84TKK101 pKa = 7.71RR102 pKa = 11.84TKK104 pKa = 9.69RR105 pKa = 11.84RR106 pKa = 11.84SSRR109 pKa = 11.84RR110 pKa = 11.84TKK112 pKa = 9.97RR113 pKa = 11.84RR114 pKa = 11.84SSRR117 pKa = 11.84RR118 pKa = 11.84STMKK122 pKa = 10.35RR123 pKa = 11.84PKK125 pKa = 9.54TSTMSKK131 pKa = 10.35GLKK134 pKa = 9.86KK135 pKa = 10.63YY136 pKa = 10.42IRR138 pKa = 11.84TKK140 pKa = 10.44SGKK143 pKa = 9.72KK144 pKa = 9.61NLSKK148 pKa = 11.04VSTKK152 pKa = 7.7MLRR155 pKa = 11.84KK156 pKa = 9.62LVALWRR162 pKa = 11.84RR163 pKa = 11.84MSAKK167 pKa = 10.19SKK169 pKa = 10.43AKK171 pKa = 9.76YY172 pKa = 9.51SVRR175 pKa = 11.84STRR178 pKa = 11.84KK179 pKa = 7.96RR180 pKa = 11.84STRR183 pKa = 11.84KK184 pKa = 8.83RR185 pKa = 11.84STRR188 pKa = 11.84KK189 pKa = 8.83RR190 pKa = 11.84STRR193 pKa = 11.84KK194 pKa = 8.83RR195 pKa = 11.84STRR198 pKa = 11.84KK199 pKa = 8.83RR200 pKa = 11.84STRR203 pKa = 11.84KK204 pKa = 8.83RR205 pKa = 11.84STRR208 pKa = 11.84KK209 pKa = 8.83RR210 pKa = 11.84STRR213 pKa = 11.84KK214 pKa = 8.83RR215 pKa = 11.84STRR218 pKa = 11.84KK219 pKa = 8.83RR220 pKa = 11.84STRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84TTRR230 pKa = 11.84KK231 pKa = 9.12RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84SRR236 pKa = 11.84PRR238 pKa = 11.84TRR240 pKa = 11.84RR241 pKa = 11.84VRR243 pKa = 11.84RR244 pKa = 11.84TRR246 pKa = 11.84RR247 pKa = 11.84TRR249 pKa = 11.84RR250 pKa = 11.84TVRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 3.12
MM1 pKa = 7.25AVSRR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84YY9 pKa = 7.36GTVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TVRR20 pKa = 11.84KK21 pKa = 8.12STASRR26 pKa = 11.84STKK29 pKa = 9.49VGRR32 pKa = 11.84VAKK35 pKa = 9.99LSNMSAGCRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.55VKK48 pKa = 10.54SQLGNRR54 pKa = 11.84KK55 pKa = 9.43LSRR58 pKa = 11.84ATRR61 pKa = 11.84ASIIRR66 pKa = 11.84RR67 pKa = 11.84SVKK70 pKa = 8.22TWSRR74 pKa = 11.84MSKK77 pKa = 9.88ARR79 pKa = 11.84KK80 pKa = 8.16SRR82 pKa = 11.84YY83 pKa = 5.01MTRR86 pKa = 11.84KK87 pKa = 8.7SSKK90 pKa = 10.01SKK92 pKa = 10.52RR93 pKa = 11.84KK94 pKa = 4.15TTRR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84TKK101 pKa = 7.71RR102 pKa = 11.84TKK104 pKa = 9.69RR105 pKa = 11.84RR106 pKa = 11.84SSRR109 pKa = 11.84RR110 pKa = 11.84TKK112 pKa = 9.97RR113 pKa = 11.84RR114 pKa = 11.84SSRR117 pKa = 11.84RR118 pKa = 11.84STMKK122 pKa = 10.35RR123 pKa = 11.84PKK125 pKa = 9.54TSTMSKK131 pKa = 10.35GLKK134 pKa = 9.86KK135 pKa = 10.63YY136 pKa = 10.42IRR138 pKa = 11.84TKK140 pKa = 10.44SGKK143 pKa = 9.72KK144 pKa = 9.61NLSKK148 pKa = 11.04VSTKK152 pKa = 7.7MLRR155 pKa = 11.84KK156 pKa = 9.62LVALWRR162 pKa = 11.84RR163 pKa = 11.84MSAKK167 pKa = 10.19SKK169 pKa = 10.43AKK171 pKa = 9.76YY172 pKa = 9.51SVRR175 pKa = 11.84STRR178 pKa = 11.84KK179 pKa = 7.96RR180 pKa = 11.84STRR183 pKa = 11.84KK184 pKa = 8.83RR185 pKa = 11.84STRR188 pKa = 11.84KK189 pKa = 8.83RR190 pKa = 11.84STRR193 pKa = 11.84KK194 pKa = 8.83RR195 pKa = 11.84STRR198 pKa = 11.84KK199 pKa = 8.83RR200 pKa = 11.84STRR203 pKa = 11.84KK204 pKa = 8.83RR205 pKa = 11.84STRR208 pKa = 11.84KK209 pKa = 8.83RR210 pKa = 11.84STRR213 pKa = 11.84KK214 pKa = 8.83RR215 pKa = 11.84STRR218 pKa = 11.84KK219 pKa = 8.83RR220 pKa = 11.84STRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84TTRR230 pKa = 11.84KK231 pKa = 9.12RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84SRR236 pKa = 11.84PRR238 pKa = 11.84TRR240 pKa = 11.84RR241 pKa = 11.84VRR243 pKa = 11.84RR244 pKa = 11.84TRR246 pKa = 11.84RR247 pKa = 11.84TRR249 pKa = 11.84RR250 pKa = 11.84TVRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 3.12
Molecular weight: 31.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
123003 |
20 |
2873 |
260.6 |
29.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.393 ± 0.123 | 1.829 ± 0.075 |
5.716 ± 0.092 | 5.094 ± 0.117 |
3.833 ± 0.083 | 5.254 ± 0.128 |
2.532 ± 0.082 | 7.259 ± 0.138 |
5.768 ± 0.204 | 7.462 ± 0.116 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.069 ± 0.092 | 5.553 ± 0.099 |
6.481 ± 0.718 | 3.018 ± 0.07 |
4.704 ± 0.119 | 7.493 ± 0.166 |
6.806 ± 0.095 | 6.416 ± 0.109 |
1.092 ± 0.041 | 4.228 ± 0.095 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
