
St Croix River virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
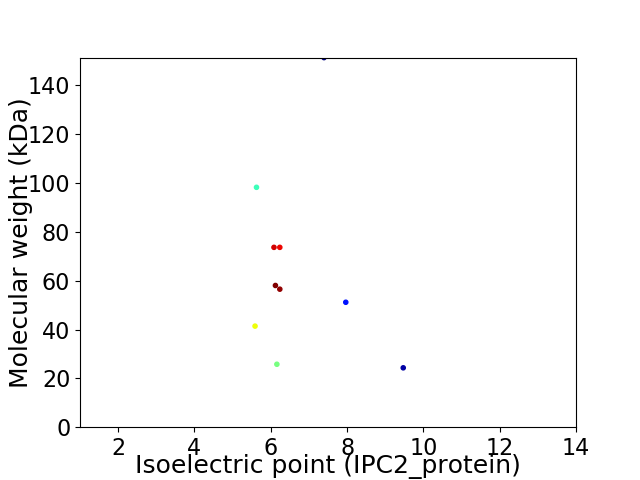
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9DSP3|Q9DSP3_9REOV NS2 OS=St Croix River virus OX=104581 PE=4 SV=1
MM1 pKa = 7.6SDD3 pKa = 2.72SRR5 pKa = 11.84AYY7 pKa = 10.33AIEE10 pKa = 4.07ILDD13 pKa = 4.04LFADD17 pKa = 3.92QVPPLPGGDD26 pKa = 3.32SRR28 pKa = 11.84ALSRR32 pKa = 11.84ARR34 pKa = 11.84AFLRR38 pKa = 11.84NPIVVRR44 pKa = 11.84NYY46 pKa = 10.54PNLDD50 pKa = 3.44PLGNDD55 pKa = 2.66ATVRR59 pKa = 11.84RR60 pKa = 11.84NLFFVCLDD68 pKa = 3.47VAMLALDD75 pKa = 3.26INEE78 pKa = 3.91FKK80 pKa = 11.15VRR82 pKa = 11.84IEE84 pKa = 3.76SHH86 pKa = 5.25RR87 pKa = 11.84VARR90 pKa = 11.84EE91 pKa = 3.49LAALLGPTGADD102 pKa = 3.43PQPTAVQIAEE112 pKa = 4.19SFTDD116 pKa = 3.29KK117 pKa = 11.15GRR119 pKa = 11.84TTYY122 pKa = 11.18AVGQDD127 pKa = 3.22GPVYY131 pKa = 10.49IEE133 pKa = 4.33GAYY136 pKa = 10.36SPSPFTYY143 pKa = 9.97GPRR146 pKa = 11.84YY147 pKa = 8.6VQDD150 pKa = 3.49GNFVNVVGPVRR161 pKa = 11.84DD162 pKa = 3.74GAAPPEE168 pKa = 4.28PCFLVANSPGRR179 pKa = 11.84VTVILNGGPNSLALEE194 pKa = 4.61HH195 pKa = 6.81IFRR198 pKa = 11.84TILRR202 pKa = 11.84RR203 pKa = 11.84GSLFTARR210 pKa = 11.84WYY212 pKa = 10.92ANSCFIGEE220 pKa = 4.15NGAPIITNGVPSLTSGGGGTNIPAYY245 pKa = 10.58FPFTFSKK252 pKa = 10.64RR253 pKa = 11.84STFTARR259 pKa = 11.84VRR261 pKa = 11.84GGGAWPVAAITIEE274 pKa = 3.93FLSFYY279 pKa = 10.55SIGHH283 pKa = 6.48FGDD286 pKa = 4.82PYY288 pKa = 10.76PGFSTDD294 pKa = 3.37LQDD297 pKa = 4.89FYY299 pKa = 11.11TYY301 pKa = 10.64TNPVWHH307 pKa = 7.62ALRR310 pKa = 11.84KK311 pKa = 8.86QLSSAMDD318 pKa = 3.96LSMYY322 pKa = 10.51VATKK326 pKa = 9.95YY327 pKa = 11.23DD328 pKa = 3.73DD329 pKa = 3.79FTRR332 pKa = 11.84HH333 pKa = 5.59QMLALALFGRR343 pKa = 11.84LSRR346 pKa = 11.84IFRR349 pKa = 11.84RR350 pKa = 11.84LDD352 pKa = 3.24PDD354 pKa = 4.28LRR356 pKa = 11.84VLPLDD361 pKa = 4.11NGDD364 pKa = 4.36GEE366 pKa = 4.63AEE368 pKa = 4.1AYY370 pKa = 9.86AAEE373 pKa = 4.33AAVEE377 pKa = 4.17EE378 pKa = 4.57AA379 pKa = 4.7
MM1 pKa = 7.6SDD3 pKa = 2.72SRR5 pKa = 11.84AYY7 pKa = 10.33AIEE10 pKa = 4.07ILDD13 pKa = 4.04LFADD17 pKa = 3.92QVPPLPGGDD26 pKa = 3.32SRR28 pKa = 11.84ALSRR32 pKa = 11.84ARR34 pKa = 11.84AFLRR38 pKa = 11.84NPIVVRR44 pKa = 11.84NYY46 pKa = 10.54PNLDD50 pKa = 3.44PLGNDD55 pKa = 2.66ATVRR59 pKa = 11.84RR60 pKa = 11.84NLFFVCLDD68 pKa = 3.47VAMLALDD75 pKa = 3.26INEE78 pKa = 3.91FKK80 pKa = 11.15VRR82 pKa = 11.84IEE84 pKa = 3.76SHH86 pKa = 5.25RR87 pKa = 11.84VARR90 pKa = 11.84EE91 pKa = 3.49LAALLGPTGADD102 pKa = 3.43PQPTAVQIAEE112 pKa = 4.19SFTDD116 pKa = 3.29KK117 pKa = 11.15GRR119 pKa = 11.84TTYY122 pKa = 11.18AVGQDD127 pKa = 3.22GPVYY131 pKa = 10.49IEE133 pKa = 4.33GAYY136 pKa = 10.36SPSPFTYY143 pKa = 9.97GPRR146 pKa = 11.84YY147 pKa = 8.6VQDD150 pKa = 3.49GNFVNVVGPVRR161 pKa = 11.84DD162 pKa = 3.74GAAPPEE168 pKa = 4.28PCFLVANSPGRR179 pKa = 11.84VTVILNGGPNSLALEE194 pKa = 4.61HH195 pKa = 6.81IFRR198 pKa = 11.84TILRR202 pKa = 11.84RR203 pKa = 11.84GSLFTARR210 pKa = 11.84WYY212 pKa = 10.92ANSCFIGEE220 pKa = 4.15NGAPIITNGVPSLTSGGGGTNIPAYY245 pKa = 10.58FPFTFSKK252 pKa = 10.64RR253 pKa = 11.84STFTARR259 pKa = 11.84VRR261 pKa = 11.84GGGAWPVAAITIEE274 pKa = 3.93FLSFYY279 pKa = 10.55SIGHH283 pKa = 6.48FGDD286 pKa = 4.82PYY288 pKa = 10.76PGFSTDD294 pKa = 3.37LQDD297 pKa = 4.89FYY299 pKa = 11.11TYY301 pKa = 10.64TNPVWHH307 pKa = 7.62ALRR310 pKa = 11.84KK311 pKa = 8.86QLSSAMDD318 pKa = 3.96LSMYY322 pKa = 10.51VATKK326 pKa = 9.95YY327 pKa = 11.23DD328 pKa = 3.73DD329 pKa = 3.79FTRR332 pKa = 11.84HH333 pKa = 5.59QMLALALFGRR343 pKa = 11.84LSRR346 pKa = 11.84IFRR349 pKa = 11.84RR350 pKa = 11.84LDD352 pKa = 3.24PDD354 pKa = 4.28LRR356 pKa = 11.84VLPLDD361 pKa = 4.11NGDD364 pKa = 4.36GEE366 pKa = 4.63AEE368 pKa = 4.1AYY370 pKa = 9.86AAEE373 pKa = 4.33AAVEE377 pKa = 4.17EE378 pKa = 4.57AA379 pKa = 4.7
Molecular weight: 41.39 kDa
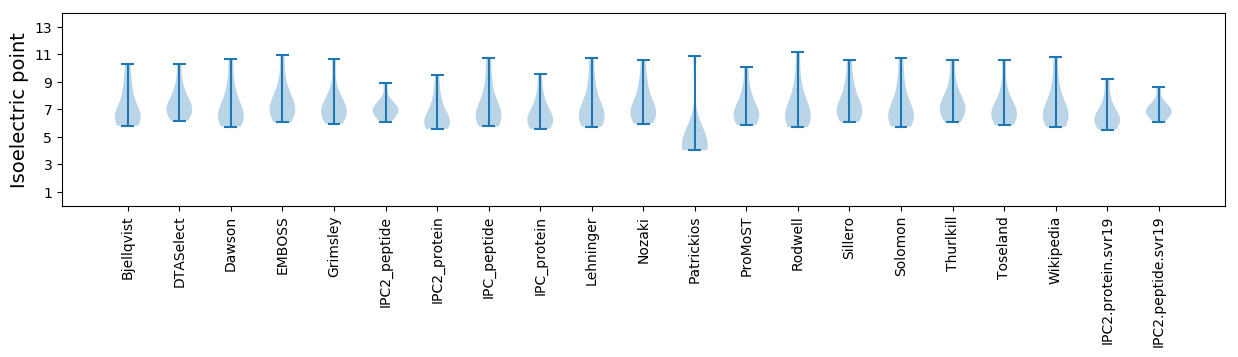
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9DSP1|Q9DSP1_9REOV VP6 OS=St Croix River virus OX=104581 PE=4 SV=1
MM1 pKa = 7.31QKK3 pKa = 10.03QSHH6 pKa = 5.82LTMMPRR12 pKa = 11.84PTAVVMTKK20 pKa = 10.64DD21 pKa = 3.38GIGAEE26 pKa = 4.36RR27 pKa = 11.84VTLMPSAPSVSPLQVISEE45 pKa = 4.41TLSVPSAKK53 pKa = 9.66TPPEE57 pKa = 4.0RR58 pKa = 11.84MEE60 pKa = 3.89KK61 pKa = 10.44AALKK65 pKa = 10.0GAQEE69 pKa = 4.91AISDD73 pKa = 3.86SSDD76 pKa = 3.89PIGMLIKK83 pKa = 10.63VEE85 pKa = 4.59ANASTISALQHH96 pKa = 5.67GEE98 pKa = 3.82RR99 pKa = 11.84KK100 pKa = 9.1LARR103 pKa = 11.84RR104 pKa = 11.84KK105 pKa = 10.15SLLFWLHH112 pKa = 5.73LTTVILMLVFTAISSIANLSIHH134 pKa = 6.53LEE136 pKa = 4.09HH137 pKa = 6.9YY138 pKa = 10.94LRR140 pKa = 11.84MANLSSNMICLGFATSTMFITRR162 pKa = 11.84TRR164 pKa = 11.84LSVALEE170 pKa = 3.64LHH172 pKa = 6.19KK173 pKa = 10.83VRR175 pKa = 11.84KK176 pKa = 7.87QLRR179 pKa = 11.84KK180 pKa = 9.45RR181 pKa = 11.84LAYY184 pKa = 9.89QATAKK189 pKa = 10.52NISAARR195 pKa = 11.84APPPSQPFLSDD206 pKa = 3.31VSSGTSGWIVAPGCKK221 pKa = 9.72ALQQ224 pKa = 3.55
MM1 pKa = 7.31QKK3 pKa = 10.03QSHH6 pKa = 5.82LTMMPRR12 pKa = 11.84PTAVVMTKK20 pKa = 10.64DD21 pKa = 3.38GIGAEE26 pKa = 4.36RR27 pKa = 11.84VTLMPSAPSVSPLQVISEE45 pKa = 4.41TLSVPSAKK53 pKa = 9.66TPPEE57 pKa = 4.0RR58 pKa = 11.84MEE60 pKa = 3.89KK61 pKa = 10.44AALKK65 pKa = 10.0GAQEE69 pKa = 4.91AISDD73 pKa = 3.86SSDD76 pKa = 3.89PIGMLIKK83 pKa = 10.63VEE85 pKa = 4.59ANASTISALQHH96 pKa = 5.67GEE98 pKa = 3.82RR99 pKa = 11.84KK100 pKa = 9.1LARR103 pKa = 11.84RR104 pKa = 11.84KK105 pKa = 10.15SLLFWLHH112 pKa = 5.73LTTVILMLVFTAISSIANLSIHH134 pKa = 6.53LEE136 pKa = 4.09HH137 pKa = 6.9YY138 pKa = 10.94LRR140 pKa = 11.84MANLSSNMICLGFATSTMFITRR162 pKa = 11.84TRR164 pKa = 11.84LSVALEE170 pKa = 3.64LHH172 pKa = 6.19KK173 pKa = 10.83VRR175 pKa = 11.84KK176 pKa = 7.87QLRR179 pKa = 11.84KK180 pKa = 9.45RR181 pKa = 11.84LAYY184 pKa = 9.89QATAKK189 pKa = 10.52NISAARR195 pKa = 11.84APPPSQPFLSDD206 pKa = 3.31VSSGTSGWIVAPGCKK221 pKa = 9.72ALQQ224 pKa = 3.55
Molecular weight: 24.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5863 |
224 |
1345 |
586.3 |
65.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.846 ± 0.771 | 0.955 ± 0.151 |
5.611 ± 0.381 | 5.151 ± 0.41 |
4.844 ± 0.364 | 5.987 ± 0.251 |
3.087 ± 0.271 | 5.373 ± 0.35 |
3.275 ± 0.318 | 10.387 ± 0.362 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.354 ± 0.193 | 2.729 ± 0.247 |
6.55 ± 0.448 | 3.377 ± 0.297 |
7.266 ± 0.315 | 9.006 ± 0.442 |
5.526 ± 0.448 | 6.669 ± 0.24 |
0.972 ± 0.222 | 3.036 ± 0.2 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
