
Common bottlenose dolphin gammaherpesvirus 1 strain Sarasota
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus; unclassified Rhadinovirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
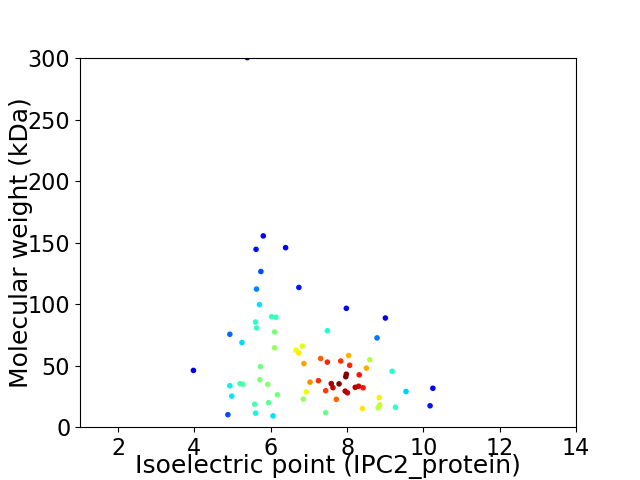
Virtual 2D-PAGE plot for 72 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
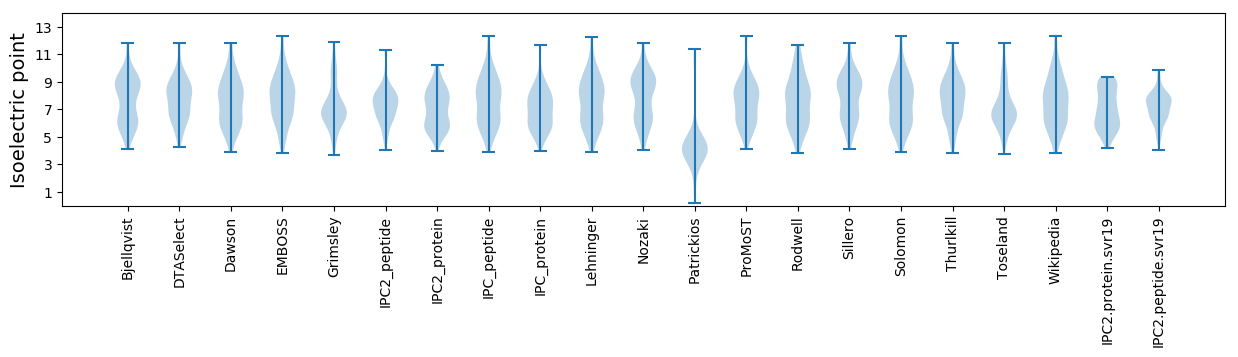
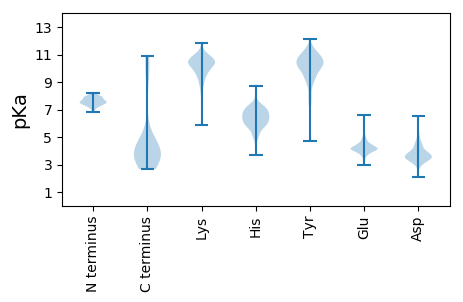
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z1NE30|A0A1Z1NE30_9GAMA DNA replication helicase OS=Common bottlenose dolphin gammaherpesvirus 1 strain Sarasota OX=2022783 GN=ORF44 PE=3 SV=1
MM1 pKa = 8.0AMFLLRR7 pKa = 11.84EE8 pKa = 4.3AKK10 pKa = 9.36VARR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PDD19 pKa = 3.2DD20 pKa = 4.05RR21 pKa = 11.84MLPIYY26 pKa = 8.97GAPRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 5.9TTEE35 pKa = 3.65YY36 pKa = 9.32FTFPTHH42 pKa = 7.32LSSLDD47 pKa = 3.8LSDD50 pKa = 4.71SSSLSPSSPLTEE62 pKa = 4.54DD63 pKa = 4.68RR64 pKa = 11.84EE65 pKa = 4.55LCSTAVTSSTLNASVGEE82 pKa = 4.09NGPCRR87 pKa = 11.84LRR89 pKa = 11.84LRR91 pKa = 11.84NLKK94 pKa = 9.51TSEE97 pKa = 3.86AALKK101 pKa = 10.53GGRR104 pKa = 11.84CSMGCGKK111 pKa = 10.11LAYY114 pKa = 9.97QRR116 pKa = 11.84IRR118 pKa = 11.84PLNSKK123 pKa = 9.63IDD125 pKa = 3.7EE126 pKa = 4.58DD127 pKa = 4.47SEE129 pKa = 4.69CEE131 pKa = 3.93EE132 pKa = 5.25GEE134 pKa = 4.49GCGGNAAASALRR146 pKa = 11.84DD147 pKa = 3.74DD148 pKa = 4.67VKK150 pKa = 11.3DD151 pKa = 3.76EE152 pKa = 4.15EE153 pKa = 5.89GEE155 pKa = 4.87DD156 pKa = 4.08GDD158 pKa = 4.88CVEE161 pKa = 5.79GEE163 pKa = 5.0DD164 pKa = 5.07GDD166 pKa = 4.98EE167 pKa = 4.43SDD169 pKa = 4.25SDD171 pKa = 4.39DD172 pKa = 4.08YY173 pKa = 12.07EE174 pKa = 4.27EE175 pKa = 6.01DD176 pKa = 3.78EE177 pKa = 4.45EE178 pKa = 5.59DD179 pKa = 4.53EE180 pKa = 4.13EE181 pKa = 5.63DD182 pKa = 4.67EE183 pKa = 4.24EE184 pKa = 5.63DD185 pKa = 4.67EE186 pKa = 4.6EE187 pKa = 5.63DD188 pKa = 5.23EE189 pKa = 4.3EE190 pKa = 6.34DD191 pKa = 3.56EE192 pKa = 5.2DD193 pKa = 4.63EE194 pKa = 4.43EE195 pKa = 5.33GEE197 pKa = 4.12EE198 pKa = 4.52GEE200 pKa = 4.42EE201 pKa = 4.74DD202 pKa = 4.81EE203 pKa = 5.37EE204 pKa = 6.36DD205 pKa = 3.72EE206 pKa = 5.76DD207 pKa = 5.63DD208 pKa = 5.28EE209 pKa = 4.89EE210 pKa = 6.49DD211 pKa = 3.97EE212 pKa = 4.59EE213 pKa = 6.1SDD215 pKa = 5.54DD216 pKa = 5.38GSEE219 pKa = 4.17LCPGDD224 pKa = 3.7SRR226 pKa = 11.84YY227 pKa = 10.64EE228 pKa = 4.01DD229 pKa = 3.61VSDD232 pKa = 4.55EE233 pKa = 4.55DD234 pKa = 4.69EE235 pKa = 4.38SDD237 pKa = 3.94CFIDD241 pKa = 4.71EE242 pKa = 4.1ADD244 pKa = 3.57KK245 pKa = 11.48VNEE248 pKa = 3.83IRR250 pKa = 11.84VADD253 pKa = 4.11EE254 pKa = 5.64DD255 pKa = 3.75IGSSDD260 pKa = 3.86TEE262 pKa = 4.56SEE264 pKa = 4.39CSSDD268 pKa = 4.72DD269 pKa = 3.69NSPCSSVSRR278 pKa = 11.84NGDD281 pKa = 3.31EE282 pKa = 4.34GDD284 pKa = 3.93VIVISSDD291 pKa = 4.04DD292 pKa = 3.65EE293 pKa = 4.84GEE295 pKa = 4.18SCLRR299 pKa = 11.84KK300 pKa = 9.62AAGDD304 pKa = 3.7GLEE307 pKa = 5.12GSDD310 pKa = 5.19SDD312 pKa = 4.47DD313 pKa = 3.87DD314 pKa = 4.23CVLVSSTYY322 pKa = 10.37KK323 pKa = 10.26RR324 pKa = 11.84RR325 pKa = 11.84LSSSSEE331 pKa = 3.79PQTPDD336 pKa = 2.66ILKK339 pKa = 9.97KK340 pKa = 7.67SARR343 pKa = 11.84MIVYY347 pKa = 10.07SSSSSDD353 pKa = 3.59EE354 pKa = 4.13EE355 pKa = 5.13DD356 pKa = 3.74DD357 pKa = 4.73SGTGSQSSVDD367 pKa = 3.58GSRR370 pKa = 11.84EE371 pKa = 3.78TGTSYY376 pKa = 11.34RR377 pKa = 11.84MPTSYY382 pKa = 11.03LPTRR386 pKa = 11.84PRR388 pKa = 11.84SCKK391 pKa = 9.86KK392 pKa = 9.92SSNRR396 pKa = 11.84YY397 pKa = 7.31SRR399 pKa = 11.84RR400 pKa = 11.84VMHH403 pKa = 6.27KK404 pKa = 7.91TPPLKK409 pKa = 10.89GHH411 pKa = 7.33DD412 pKa = 3.98CYY414 pKa = 11.5NWPWLDD420 pKa = 3.21
MM1 pKa = 8.0AMFLLRR7 pKa = 11.84EE8 pKa = 4.3AKK10 pKa = 9.36VARR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PDD19 pKa = 3.2DD20 pKa = 4.05RR21 pKa = 11.84MLPIYY26 pKa = 8.97GAPRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 5.9TTEE35 pKa = 3.65YY36 pKa = 9.32FTFPTHH42 pKa = 7.32LSSLDD47 pKa = 3.8LSDD50 pKa = 4.71SSSLSPSSPLTEE62 pKa = 4.54DD63 pKa = 4.68RR64 pKa = 11.84EE65 pKa = 4.55LCSTAVTSSTLNASVGEE82 pKa = 4.09NGPCRR87 pKa = 11.84LRR89 pKa = 11.84LRR91 pKa = 11.84NLKK94 pKa = 9.51TSEE97 pKa = 3.86AALKK101 pKa = 10.53GGRR104 pKa = 11.84CSMGCGKK111 pKa = 10.11LAYY114 pKa = 9.97QRR116 pKa = 11.84IRR118 pKa = 11.84PLNSKK123 pKa = 9.63IDD125 pKa = 3.7EE126 pKa = 4.58DD127 pKa = 4.47SEE129 pKa = 4.69CEE131 pKa = 3.93EE132 pKa = 5.25GEE134 pKa = 4.49GCGGNAAASALRR146 pKa = 11.84DD147 pKa = 3.74DD148 pKa = 4.67VKK150 pKa = 11.3DD151 pKa = 3.76EE152 pKa = 4.15EE153 pKa = 5.89GEE155 pKa = 4.87DD156 pKa = 4.08GDD158 pKa = 4.88CVEE161 pKa = 5.79GEE163 pKa = 5.0DD164 pKa = 5.07GDD166 pKa = 4.98EE167 pKa = 4.43SDD169 pKa = 4.25SDD171 pKa = 4.39DD172 pKa = 4.08YY173 pKa = 12.07EE174 pKa = 4.27EE175 pKa = 6.01DD176 pKa = 3.78EE177 pKa = 4.45EE178 pKa = 5.59DD179 pKa = 4.53EE180 pKa = 4.13EE181 pKa = 5.63DD182 pKa = 4.67EE183 pKa = 4.24EE184 pKa = 5.63DD185 pKa = 4.67EE186 pKa = 4.6EE187 pKa = 5.63DD188 pKa = 5.23EE189 pKa = 4.3EE190 pKa = 6.34DD191 pKa = 3.56EE192 pKa = 5.2DD193 pKa = 4.63EE194 pKa = 4.43EE195 pKa = 5.33GEE197 pKa = 4.12EE198 pKa = 4.52GEE200 pKa = 4.42EE201 pKa = 4.74DD202 pKa = 4.81EE203 pKa = 5.37EE204 pKa = 6.36DD205 pKa = 3.72EE206 pKa = 5.76DD207 pKa = 5.63DD208 pKa = 5.28EE209 pKa = 4.89EE210 pKa = 6.49DD211 pKa = 3.97EE212 pKa = 4.59EE213 pKa = 6.1SDD215 pKa = 5.54DD216 pKa = 5.38GSEE219 pKa = 4.17LCPGDD224 pKa = 3.7SRR226 pKa = 11.84YY227 pKa = 10.64EE228 pKa = 4.01DD229 pKa = 3.61VSDD232 pKa = 4.55EE233 pKa = 4.55DD234 pKa = 4.69EE235 pKa = 4.38SDD237 pKa = 3.94CFIDD241 pKa = 4.71EE242 pKa = 4.1ADD244 pKa = 3.57KK245 pKa = 11.48VNEE248 pKa = 3.83IRR250 pKa = 11.84VADD253 pKa = 4.11EE254 pKa = 5.64DD255 pKa = 3.75IGSSDD260 pKa = 3.86TEE262 pKa = 4.56SEE264 pKa = 4.39CSSDD268 pKa = 4.72DD269 pKa = 3.69NSPCSSVSRR278 pKa = 11.84NGDD281 pKa = 3.31EE282 pKa = 4.34GDD284 pKa = 3.93VIVISSDD291 pKa = 4.04DD292 pKa = 3.65EE293 pKa = 4.84GEE295 pKa = 4.18SCLRR299 pKa = 11.84KK300 pKa = 9.62AAGDD304 pKa = 3.7GLEE307 pKa = 5.12GSDD310 pKa = 5.19SDD312 pKa = 4.47DD313 pKa = 3.87DD314 pKa = 4.23CVLVSSTYY322 pKa = 10.37KK323 pKa = 10.26RR324 pKa = 11.84RR325 pKa = 11.84LSSSSEE331 pKa = 3.79PQTPDD336 pKa = 2.66ILKK339 pKa = 9.97KK340 pKa = 7.67SARR343 pKa = 11.84MIVYY347 pKa = 10.07SSSSSDD353 pKa = 3.59EE354 pKa = 4.13EE355 pKa = 5.13DD356 pKa = 3.74DD357 pKa = 4.73SGTGSQSSVDD367 pKa = 3.58GSRR370 pKa = 11.84EE371 pKa = 3.78TGTSYY376 pKa = 11.34RR377 pKa = 11.84MPTSYY382 pKa = 11.03LPTRR386 pKa = 11.84PRR388 pKa = 11.84SCKK391 pKa = 9.86KK392 pKa = 9.92SSNRR396 pKa = 11.84YY397 pKa = 7.31SRR399 pKa = 11.84RR400 pKa = 11.84VMHH403 pKa = 6.27KK404 pKa = 7.91TPPLKK409 pKa = 10.89GHH411 pKa = 7.33DD412 pKa = 3.98CYY414 pKa = 11.5NWPWLDD420 pKa = 3.21
Molecular weight: 46.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z1NEG6|A0A1Z1NEG6_9GAMA Shutoff alkaline exonuclease OS=Common bottlenose dolphin gammaherpesvirus 1 strain Sarasota OX=2022783 GN=ORF37 PE=3 SV=1
MM1 pKa = 7.48SGGVIPGLGDD11 pKa = 3.39ANAVLGTLPARR22 pKa = 11.84CKK24 pKa = 10.36AAGKK28 pKa = 9.42KK29 pKa = 9.82AHH31 pKa = 6.39LKK33 pKa = 10.3VYY35 pKa = 10.5RR36 pKa = 11.84KK37 pKa = 10.09LLGFKK42 pKa = 7.87TLRR45 pKa = 11.84GLSLFLDD52 pKa = 4.17LGQPVEE58 pKa = 4.63SSAKK62 pKa = 9.26HH63 pKa = 5.65RR64 pKa = 11.84IFFEE68 pKa = 3.88VRR70 pKa = 11.84LGRR73 pKa = 11.84RR74 pKa = 11.84IADD77 pKa = 3.38CVLIVKK83 pKa = 9.12TEE85 pKa = 3.87DD86 pKa = 3.03RR87 pKa = 11.84GVCYY91 pKa = 10.07IIEE94 pKa = 5.1LKK96 pKa = 9.68TCMTQNVNIFSPIRR110 pKa = 11.84QAQRR114 pKa = 11.84LQGLAQLSDD123 pKa = 3.49SVKK126 pKa = 10.21FLRR129 pKa = 11.84DD130 pKa = 3.55ALPSGPEE137 pKa = 3.47SWFIIPVLLFKK148 pKa = 11.02SQGSFKK154 pKa = 9.62TLHH157 pKa = 6.12SEE159 pKa = 5.06TYY161 pKa = 9.52QSSSQGARR169 pKa = 11.84TSWEE173 pKa = 3.72RR174 pKa = 11.84LTNFLFVRR182 pKa = 11.84EE183 pKa = 4.21DD184 pKa = 3.71DD185 pKa = 4.1AVTGALRR192 pKa = 11.84QNRR195 pKa = 11.84RR196 pKa = 11.84PRR198 pKa = 11.84VVPDD202 pKa = 3.58RR203 pKa = 11.84NLLGPQVVKK212 pKa = 10.18RR213 pKa = 11.84HH214 pKa = 4.09VRR216 pKa = 11.84KK217 pKa = 9.71RR218 pKa = 11.84RR219 pKa = 11.84PPQRR223 pKa = 11.84GDD225 pKa = 3.2APGAPTARR233 pKa = 11.84TQRR236 pKa = 11.84GPRR239 pKa = 11.84GTGGDD244 pKa = 4.19CEE246 pKa = 5.5LSVQGAAPAGHH257 pKa = 7.19DD258 pKa = 3.7EE259 pKa = 4.23ARR261 pKa = 11.84GVQQ264 pKa = 3.31
MM1 pKa = 7.48SGGVIPGLGDD11 pKa = 3.39ANAVLGTLPARR22 pKa = 11.84CKK24 pKa = 10.36AAGKK28 pKa = 9.42KK29 pKa = 9.82AHH31 pKa = 6.39LKK33 pKa = 10.3VYY35 pKa = 10.5RR36 pKa = 11.84KK37 pKa = 10.09LLGFKK42 pKa = 7.87TLRR45 pKa = 11.84GLSLFLDD52 pKa = 4.17LGQPVEE58 pKa = 4.63SSAKK62 pKa = 9.26HH63 pKa = 5.65RR64 pKa = 11.84IFFEE68 pKa = 3.88VRR70 pKa = 11.84LGRR73 pKa = 11.84RR74 pKa = 11.84IADD77 pKa = 3.38CVLIVKK83 pKa = 9.12TEE85 pKa = 3.87DD86 pKa = 3.03RR87 pKa = 11.84GVCYY91 pKa = 10.07IIEE94 pKa = 5.1LKK96 pKa = 9.68TCMTQNVNIFSPIRR110 pKa = 11.84QAQRR114 pKa = 11.84LQGLAQLSDD123 pKa = 3.49SVKK126 pKa = 10.21FLRR129 pKa = 11.84DD130 pKa = 3.55ALPSGPEE137 pKa = 3.47SWFIIPVLLFKK148 pKa = 11.02SQGSFKK154 pKa = 9.62TLHH157 pKa = 6.12SEE159 pKa = 5.06TYY161 pKa = 9.52QSSSQGARR169 pKa = 11.84TSWEE173 pKa = 3.72RR174 pKa = 11.84LTNFLFVRR182 pKa = 11.84EE183 pKa = 4.21DD184 pKa = 3.71DD185 pKa = 4.1AVTGALRR192 pKa = 11.84QNRR195 pKa = 11.84RR196 pKa = 11.84PRR198 pKa = 11.84VVPDD202 pKa = 3.58RR203 pKa = 11.84NLLGPQVVKK212 pKa = 10.18RR213 pKa = 11.84HH214 pKa = 4.09VRR216 pKa = 11.84KK217 pKa = 9.71RR218 pKa = 11.84RR219 pKa = 11.84PPQRR223 pKa = 11.84GDD225 pKa = 3.2APGAPTARR233 pKa = 11.84TQRR236 pKa = 11.84GPRR239 pKa = 11.84GTGGDD244 pKa = 4.19CEE246 pKa = 5.5LSVQGAAPAGHH257 pKa = 7.19DD258 pKa = 3.7EE259 pKa = 4.23ARR261 pKa = 11.84GVQQ264 pKa = 3.31
Molecular weight: 29.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
35681 |
83 |
2739 |
495.6 |
55.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.144 ± 0.249 | 2.416 ± 0.186 |
4.997 ± 0.168 | 5.073 ± 0.211 |
4.568 ± 0.188 | 5.762 ± 0.201 |
3.105 ± 0.113 | 4.975 ± 0.185 |
4.515 ± 0.184 | 9.938 ± 0.224 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.071 ± 0.104 | 4.235 ± 0.142 |
7.135 ± 0.606 | 3.43 ± 0.128 |
5.518 ± 0.163 | 7.533 ± 0.189 |
5.908 ± 0.198 | 6.474 ± 0.236 |
1.076 ± 0.064 | 3.125 ± 0.147 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
