
Wuhan fly virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
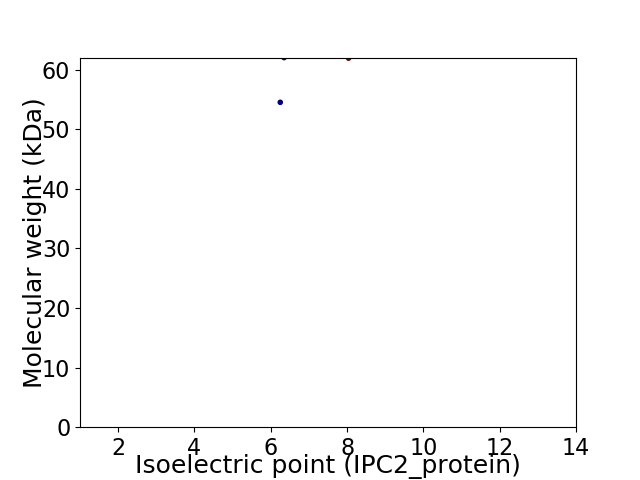
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLL7|A0A1L3KLL7_9VIRU Uncharacterized protein OS=Wuhan fly virus 5 OX=1923699 PE=4 SV=1
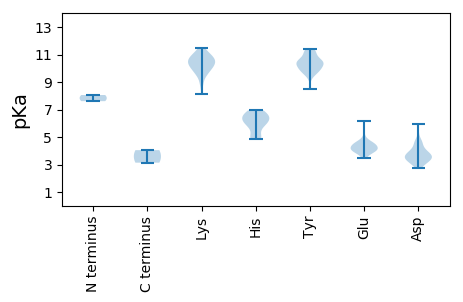
MM1 pKa = 7.61SRR3 pKa = 11.84YY4 pKa = 9.82NFNNNNNGGNFQGFVPDD21 pKa = 4.2FNNWRR26 pKa = 11.84DD27 pKa = 3.44PNMSYY32 pKa = 10.53QYY34 pKa = 11.38NNQQQQRR41 pKa = 11.84PQQNFGPQRR50 pKa = 11.84GGRR53 pKa = 11.84PHH55 pKa = 6.82VFRR58 pKa = 11.84GRR60 pKa = 11.84GGRR63 pKa = 11.84RR64 pKa = 11.84GGISNNPKK72 pKa = 10.06ANVRR76 pKa = 11.84NPVEE80 pKa = 4.48LKK82 pKa = 10.85EE83 pKa = 4.12EE84 pKa = 4.38TPKK87 pKa = 9.47PTTMPTGQHH96 pKa = 5.55LSKK99 pKa = 10.1TNSEE103 pKa = 4.54FKK105 pKa = 10.72NDD107 pKa = 4.01PLYY110 pKa = 11.31LEE112 pKa = 5.52AFMPSPSEE120 pKa = 3.71EE121 pKa = 3.94LVPIAEE127 pKa = 4.58EE128 pKa = 4.31LNHH131 pKa = 6.33FPGFDD136 pKa = 3.31GLTTLIEE143 pKa = 3.88EE144 pKa = 4.58TYY146 pKa = 11.12EE147 pKa = 3.92NFSAHH152 pKa = 5.88NLNFKK157 pKa = 10.62RR158 pKa = 11.84SVSLSAYY165 pKa = 9.57SYY167 pKa = 10.86YY168 pKa = 9.53ITVLSWARR176 pKa = 11.84VLYY179 pKa = 9.6LKK181 pKa = 10.84KK182 pKa = 9.48MNKK185 pKa = 9.51YY186 pKa = 10.45KK187 pKa = 10.14LTTNEE192 pKa = 3.98LEE194 pKa = 4.64FIEE197 pKa = 4.27MVYY200 pKa = 10.51EE201 pKa = 3.86QGNFLLPKK209 pKa = 10.21SVIIYY214 pKa = 10.45LSGFGNFNIPSGVEE228 pKa = 3.8SKK230 pKa = 11.15FNLKK234 pKa = 10.25PYY236 pKa = 10.01TYY238 pKa = 10.61DD239 pKa = 3.18EE240 pKa = 4.58LGYY243 pKa = 10.62FVDD246 pKa = 5.92FDD248 pKa = 4.16TKK250 pKa = 10.77FYY252 pKa = 11.15LCTSYY257 pKa = 11.38PNISIAAEE265 pKa = 3.5RR266 pKa = 11.84VMRR269 pKa = 11.84DD270 pKa = 3.19LAYY273 pKa = 9.83TDD275 pKa = 3.46NPEE278 pKa = 4.41IGEE281 pKa = 4.12AWSPAEE287 pKa = 6.16IEE289 pKa = 4.38QEE291 pKa = 3.84WNTRR295 pKa = 11.84CIGYY299 pKa = 9.8APSVIIHH306 pKa = 6.51DD307 pKa = 3.84MQRR310 pKa = 11.84AVLEE314 pKa = 4.22RR315 pKa = 11.84ALVRR319 pKa = 11.84SDD321 pKa = 4.55NFPSDD326 pKa = 3.79LNGFLVNVRR335 pKa = 11.84LMNNIQKK342 pKa = 9.67YY343 pKa = 9.67LSEE346 pKa = 4.32IPSLEE351 pKa = 4.09SGPIPKK357 pKa = 10.07NLTGSLGQFIIEE369 pKa = 4.24IPTVSVGANDD379 pKa = 4.28EE380 pKa = 4.2IGSLSFTSKK389 pKa = 10.96SPLSCPGSMSFLGGSFLYY407 pKa = 10.38RR408 pKa = 11.84VDD410 pKa = 4.66KK411 pKa = 11.03DD412 pKa = 3.31LTPARR417 pKa = 11.84RR418 pKa = 11.84KK419 pKa = 10.05YY420 pKa = 10.22FYY422 pKa = 9.93PYY424 pKa = 10.13TKK426 pKa = 10.68DD427 pKa = 3.53SDD429 pKa = 3.94TQEE432 pKa = 4.69DD433 pKa = 4.7KK434 pKa = 11.39INLNLLNTGWSPIFNQIYY452 pKa = 9.59HH453 pKa = 5.92YY454 pKa = 11.24SNVPFKK460 pKa = 10.73PVLRR464 pKa = 11.84VKK466 pKa = 10.05KK467 pKa = 10.18FCSIDD472 pKa = 3.54VKK474 pKa = 10.99PSGVV478 pKa = 3.13
MM1 pKa = 7.61SRR3 pKa = 11.84YY4 pKa = 9.82NFNNNNNGGNFQGFVPDD21 pKa = 4.2FNNWRR26 pKa = 11.84DD27 pKa = 3.44PNMSYY32 pKa = 10.53QYY34 pKa = 11.38NNQQQQRR41 pKa = 11.84PQQNFGPQRR50 pKa = 11.84GGRR53 pKa = 11.84PHH55 pKa = 6.82VFRR58 pKa = 11.84GRR60 pKa = 11.84GGRR63 pKa = 11.84RR64 pKa = 11.84GGISNNPKK72 pKa = 10.06ANVRR76 pKa = 11.84NPVEE80 pKa = 4.48LKK82 pKa = 10.85EE83 pKa = 4.12EE84 pKa = 4.38TPKK87 pKa = 9.47PTTMPTGQHH96 pKa = 5.55LSKK99 pKa = 10.1TNSEE103 pKa = 4.54FKK105 pKa = 10.72NDD107 pKa = 4.01PLYY110 pKa = 11.31LEE112 pKa = 5.52AFMPSPSEE120 pKa = 3.71EE121 pKa = 3.94LVPIAEE127 pKa = 4.58EE128 pKa = 4.31LNHH131 pKa = 6.33FPGFDD136 pKa = 3.31GLTTLIEE143 pKa = 3.88EE144 pKa = 4.58TYY146 pKa = 11.12EE147 pKa = 3.92NFSAHH152 pKa = 5.88NLNFKK157 pKa = 10.62RR158 pKa = 11.84SVSLSAYY165 pKa = 9.57SYY167 pKa = 10.86YY168 pKa = 9.53ITVLSWARR176 pKa = 11.84VLYY179 pKa = 9.6LKK181 pKa = 10.84KK182 pKa = 9.48MNKK185 pKa = 9.51YY186 pKa = 10.45KK187 pKa = 10.14LTTNEE192 pKa = 3.98LEE194 pKa = 4.64FIEE197 pKa = 4.27MVYY200 pKa = 10.51EE201 pKa = 3.86QGNFLLPKK209 pKa = 10.21SVIIYY214 pKa = 10.45LSGFGNFNIPSGVEE228 pKa = 3.8SKK230 pKa = 11.15FNLKK234 pKa = 10.25PYY236 pKa = 10.01TYY238 pKa = 10.61DD239 pKa = 3.18EE240 pKa = 4.58LGYY243 pKa = 10.62FVDD246 pKa = 5.92FDD248 pKa = 4.16TKK250 pKa = 10.77FYY252 pKa = 11.15LCTSYY257 pKa = 11.38PNISIAAEE265 pKa = 3.5RR266 pKa = 11.84VMRR269 pKa = 11.84DD270 pKa = 3.19LAYY273 pKa = 9.83TDD275 pKa = 3.46NPEE278 pKa = 4.41IGEE281 pKa = 4.12AWSPAEE287 pKa = 6.16IEE289 pKa = 4.38QEE291 pKa = 3.84WNTRR295 pKa = 11.84CIGYY299 pKa = 9.8APSVIIHH306 pKa = 6.51DD307 pKa = 3.84MQRR310 pKa = 11.84AVLEE314 pKa = 4.22RR315 pKa = 11.84ALVRR319 pKa = 11.84SDD321 pKa = 4.55NFPSDD326 pKa = 3.79LNGFLVNVRR335 pKa = 11.84LMNNIQKK342 pKa = 9.67YY343 pKa = 9.67LSEE346 pKa = 4.32IPSLEE351 pKa = 4.09SGPIPKK357 pKa = 10.07NLTGSLGQFIIEE369 pKa = 4.24IPTVSVGANDD379 pKa = 4.28EE380 pKa = 4.2IGSLSFTSKK389 pKa = 10.96SPLSCPGSMSFLGGSFLYY407 pKa = 10.38RR408 pKa = 11.84VDD410 pKa = 4.66KK411 pKa = 11.03DD412 pKa = 3.31LTPARR417 pKa = 11.84RR418 pKa = 11.84KK419 pKa = 10.05YY420 pKa = 10.22FYY422 pKa = 9.93PYY424 pKa = 10.13TKK426 pKa = 10.68DD427 pKa = 3.53SDD429 pKa = 3.94TQEE432 pKa = 4.69DD433 pKa = 4.7KK434 pKa = 11.39INLNLLNTGWSPIFNQIYY452 pKa = 9.59HH453 pKa = 5.92YY454 pKa = 11.24SNVPFKK460 pKa = 10.73PVLRR464 pKa = 11.84VKK466 pKa = 10.05KK467 pKa = 10.18FCSIDD472 pKa = 3.54VKK474 pKa = 10.99PSGVV478 pKa = 3.13
Molecular weight: 54.51 kDa
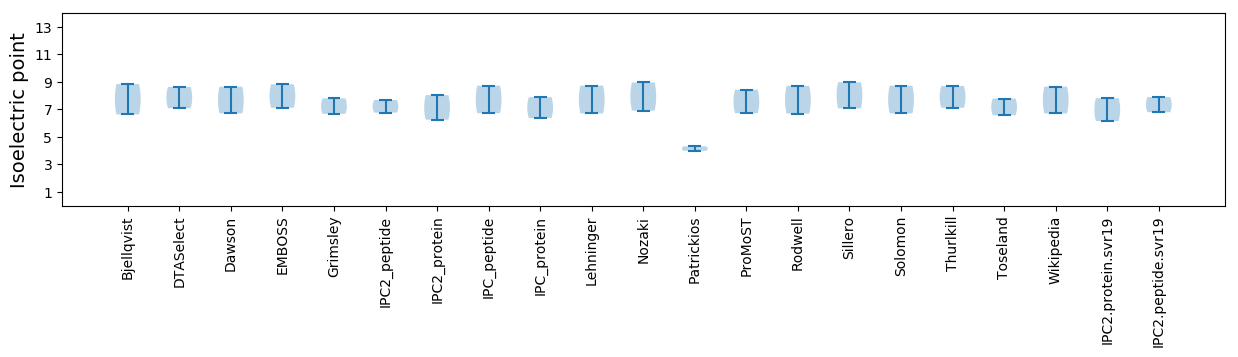
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLL7|A0A1L3KLL7_9VIRU Uncharacterized protein OS=Wuhan fly virus 5 OX=1923699 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84SRR4 pKa = 11.84GPATRR9 pKa = 11.84QCINVSPRR17 pKa = 11.84RR18 pKa = 11.84GTKK21 pKa = 8.16TAITRR26 pKa = 11.84LEE28 pKa = 4.1NKK30 pKa = 9.74EE31 pKa = 4.03GFVMNMTPARR41 pKa = 11.84EE42 pKa = 4.02DD43 pKa = 3.03GKK45 pKa = 11.15ARR47 pKa = 11.84LACEE51 pKa = 4.58LYY53 pKa = 10.28FGKK56 pKa = 10.4DD57 pKa = 2.72ITDD60 pKa = 4.72KK61 pKa = 10.35ITSTLHH67 pKa = 6.69RR68 pKa = 11.84SRR70 pKa = 11.84LTYY73 pKa = 10.7DD74 pKa = 3.24SLLEE78 pKa = 4.65DD79 pKa = 3.82FMDD82 pKa = 3.8YY83 pKa = 11.32DD84 pKa = 3.79RR85 pKa = 11.84THH87 pKa = 6.6VPRR90 pKa = 11.84ISDD93 pKa = 3.52PDD95 pKa = 3.25AQIIYY100 pKa = 10.06KK101 pKa = 10.42CVLQSIKK108 pKa = 10.87DD109 pKa = 3.51DD110 pKa = 4.01LGNVKK115 pKa = 10.48LSPLTYY121 pKa = 9.85QQVRR125 pKa = 11.84DD126 pKa = 3.89LPDD129 pKa = 3.64FPGAKK134 pKa = 9.94SPGLPYY140 pKa = 10.74KK141 pKa = 10.81NLGFTKK147 pKa = 10.45KK148 pKa = 10.41SDD150 pKa = 3.58VYY152 pKa = 11.11DD153 pKa = 3.89DD154 pKa = 4.7PIYY157 pKa = 10.5RR158 pKa = 11.84EE159 pKa = 4.05HH160 pKa = 6.94LTNLWRR166 pKa = 11.84NIGHH170 pKa = 6.73GKK172 pKa = 8.77NVKK175 pKa = 10.45LPDD178 pKa = 3.05VCMFARR184 pKa = 11.84AQVAKK189 pKa = 10.45APKK192 pKa = 9.59NKK194 pKa = 8.93IRR196 pKa = 11.84ATWGYY201 pKa = 10.4SFDD204 pKa = 3.69VYY206 pKa = 10.53MEE208 pKa = 4.19EE209 pKa = 4.97ARR211 pKa = 11.84FFYY214 pKa = 10.03PIQEE218 pKa = 4.4FIKK221 pKa = 9.9SHH223 pKa = 4.97KK224 pKa = 9.78HH225 pKa = 4.88NLPIAYY231 pKa = 9.24GLEE234 pKa = 4.01MANGGMVAINDD245 pKa = 3.29MLQRR249 pKa = 11.84HH250 pKa = 5.94RR251 pKa = 11.84NSKK254 pKa = 10.28YY255 pKa = 10.93VISDD259 pKa = 2.88WSKK262 pKa = 9.92FDD264 pKa = 3.25KK265 pKa = 10.72TIPPWLIRR273 pKa = 11.84DD274 pKa = 3.46AFEE277 pKa = 4.35ILEE280 pKa = 3.97GLIDD284 pKa = 4.31FSAVHH289 pKa = 6.53RR290 pKa = 11.84SAHH293 pKa = 4.99EE294 pKa = 3.52KK295 pKa = 10.49RR296 pKa = 11.84RR297 pKa = 11.84FKK299 pKa = 11.06KK300 pKa = 10.26LVDD303 pKa = 3.85YY304 pKa = 10.48FVEE307 pKa = 4.67TPIRR311 pKa = 11.84TCKK314 pKa = 10.63GEE316 pKa = 3.98RR317 pKa = 11.84FLVTGGVPSGSCFTNIIDD335 pKa = 5.23SIINCIVTRR344 pKa = 11.84FLVFQTTNAFPVGEE358 pKa = 4.59IFLGDD363 pKa = 3.67DD364 pKa = 3.54GVFIVDD370 pKa = 4.62GFACLEE376 pKa = 4.98DD377 pKa = 3.63IASLAIKK384 pKa = 9.83YY385 pKa = 10.19FGMILNVDD393 pKa = 3.4KK394 pKa = 11.19SYY396 pKa = 10.2VTTNPSNVHH405 pKa = 4.86FLGYY409 pKa = 10.08FNYY412 pKa = 10.41SGYY415 pKa = 9.82PFKK418 pKa = 11.38NQDD421 pKa = 3.13FLIASFIFPEE431 pKa = 4.29HH432 pKa = 6.29KK433 pKa = 9.4RR434 pKa = 11.84TRR436 pKa = 11.84LVDD439 pKa = 3.48ACAAALGQMYY449 pKa = 10.5SGFDD453 pKa = 3.05PGYY456 pKa = 10.41ARR458 pKa = 11.84NWLNIIHH465 pKa = 6.25YY466 pKa = 10.09LADD469 pKa = 4.23LEE471 pKa = 4.27NSNPFSLNEE480 pKa = 4.05VVLHH484 pKa = 6.16LKK486 pKa = 10.57NNEE489 pKa = 3.6FRR491 pKa = 11.84HH492 pKa = 6.11KK493 pKa = 10.65YY494 pKa = 8.47LAQVGISADD503 pKa = 3.56SMTLPSRR510 pKa = 11.84ANSQILEE517 pKa = 4.34VLPKK521 pKa = 9.04TYY523 pKa = 10.88CNITLPSRR531 pKa = 11.84AYY533 pKa = 10.18DD534 pKa = 3.59FKK536 pKa = 11.45DD537 pKa = 2.76ILSRR541 pKa = 11.84II542 pKa = 4.05
MM1 pKa = 8.06RR2 pKa = 11.84SRR4 pKa = 11.84GPATRR9 pKa = 11.84QCINVSPRR17 pKa = 11.84RR18 pKa = 11.84GTKK21 pKa = 8.16TAITRR26 pKa = 11.84LEE28 pKa = 4.1NKK30 pKa = 9.74EE31 pKa = 4.03GFVMNMTPARR41 pKa = 11.84EE42 pKa = 4.02DD43 pKa = 3.03GKK45 pKa = 11.15ARR47 pKa = 11.84LACEE51 pKa = 4.58LYY53 pKa = 10.28FGKK56 pKa = 10.4DD57 pKa = 2.72ITDD60 pKa = 4.72KK61 pKa = 10.35ITSTLHH67 pKa = 6.69RR68 pKa = 11.84SRR70 pKa = 11.84LTYY73 pKa = 10.7DD74 pKa = 3.24SLLEE78 pKa = 4.65DD79 pKa = 3.82FMDD82 pKa = 3.8YY83 pKa = 11.32DD84 pKa = 3.79RR85 pKa = 11.84THH87 pKa = 6.6VPRR90 pKa = 11.84ISDD93 pKa = 3.52PDD95 pKa = 3.25AQIIYY100 pKa = 10.06KK101 pKa = 10.42CVLQSIKK108 pKa = 10.87DD109 pKa = 3.51DD110 pKa = 4.01LGNVKK115 pKa = 10.48LSPLTYY121 pKa = 9.85QQVRR125 pKa = 11.84DD126 pKa = 3.89LPDD129 pKa = 3.64FPGAKK134 pKa = 9.94SPGLPYY140 pKa = 10.74KK141 pKa = 10.81NLGFTKK147 pKa = 10.45KK148 pKa = 10.41SDD150 pKa = 3.58VYY152 pKa = 11.11DD153 pKa = 3.89DD154 pKa = 4.7PIYY157 pKa = 10.5RR158 pKa = 11.84EE159 pKa = 4.05HH160 pKa = 6.94LTNLWRR166 pKa = 11.84NIGHH170 pKa = 6.73GKK172 pKa = 8.77NVKK175 pKa = 10.45LPDD178 pKa = 3.05VCMFARR184 pKa = 11.84AQVAKK189 pKa = 10.45APKK192 pKa = 9.59NKK194 pKa = 8.93IRR196 pKa = 11.84ATWGYY201 pKa = 10.4SFDD204 pKa = 3.69VYY206 pKa = 10.53MEE208 pKa = 4.19EE209 pKa = 4.97ARR211 pKa = 11.84FFYY214 pKa = 10.03PIQEE218 pKa = 4.4FIKK221 pKa = 9.9SHH223 pKa = 4.97KK224 pKa = 9.78HH225 pKa = 4.88NLPIAYY231 pKa = 9.24GLEE234 pKa = 4.01MANGGMVAINDD245 pKa = 3.29MLQRR249 pKa = 11.84HH250 pKa = 5.94RR251 pKa = 11.84NSKK254 pKa = 10.28YY255 pKa = 10.93VISDD259 pKa = 2.88WSKK262 pKa = 9.92FDD264 pKa = 3.25KK265 pKa = 10.72TIPPWLIRR273 pKa = 11.84DD274 pKa = 3.46AFEE277 pKa = 4.35ILEE280 pKa = 3.97GLIDD284 pKa = 4.31FSAVHH289 pKa = 6.53RR290 pKa = 11.84SAHH293 pKa = 4.99EE294 pKa = 3.52KK295 pKa = 10.49RR296 pKa = 11.84RR297 pKa = 11.84FKK299 pKa = 11.06KK300 pKa = 10.26LVDD303 pKa = 3.85YY304 pKa = 10.48FVEE307 pKa = 4.67TPIRR311 pKa = 11.84TCKK314 pKa = 10.63GEE316 pKa = 3.98RR317 pKa = 11.84FLVTGGVPSGSCFTNIIDD335 pKa = 5.23SIINCIVTRR344 pKa = 11.84FLVFQTTNAFPVGEE358 pKa = 4.59IFLGDD363 pKa = 3.67DD364 pKa = 3.54GVFIVDD370 pKa = 4.62GFACLEE376 pKa = 4.98DD377 pKa = 3.63IASLAIKK384 pKa = 9.83YY385 pKa = 10.19FGMILNVDD393 pKa = 3.4KK394 pKa = 11.19SYY396 pKa = 10.2VTTNPSNVHH405 pKa = 4.86FLGYY409 pKa = 10.08FNYY412 pKa = 10.41SGYY415 pKa = 9.82PFKK418 pKa = 11.38NQDD421 pKa = 3.13FLIASFIFPEE431 pKa = 4.29HH432 pKa = 6.29KK433 pKa = 9.4RR434 pKa = 11.84TRR436 pKa = 11.84LVDD439 pKa = 3.48ACAAALGQMYY449 pKa = 10.5SGFDD453 pKa = 3.05PGYY456 pKa = 10.41ARR458 pKa = 11.84NWLNIIHH465 pKa = 6.25YY466 pKa = 10.09LADD469 pKa = 4.23LEE471 pKa = 4.27NSNPFSLNEE480 pKa = 4.05VVLHH484 pKa = 6.16LKK486 pKa = 10.57NNEE489 pKa = 3.6FRR491 pKa = 11.84HH492 pKa = 6.11KK493 pKa = 10.65YY494 pKa = 8.47LAQVGISADD503 pKa = 3.56SMTLPSRR510 pKa = 11.84ANSQILEE517 pKa = 4.34VLPKK521 pKa = 9.04TYY523 pKa = 10.88CNITLPSRR531 pKa = 11.84AYY533 pKa = 10.18DD534 pKa = 3.59FKK536 pKa = 11.45DD537 pKa = 2.76ILSRR541 pKa = 11.84II542 pKa = 4.05
Molecular weight: 61.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1020 |
478 |
542 |
510.0 |
58.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.902 ± 1.026 | 1.373 ± 0.353 |
5.49 ± 1.0 | 5.098 ± 0.777 |
6.275 ± 0.001 | 6.176 ± 0.342 |
1.961 ± 0.465 | 6.471 ± 0.68 |
5.784 ± 0.366 | 8.431 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.157 ± 0.043 | 7.255 ± 1.286 |
6.176 ± 0.756 | 2.941 ± 0.406 |
5.588 ± 0.512 | 7.353 ± 0.808 |
5.0 ± 0.124 | 5.588 ± 0.098 |
0.98 ± 0.043 | 5.0 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
