
Beihai sobemo-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
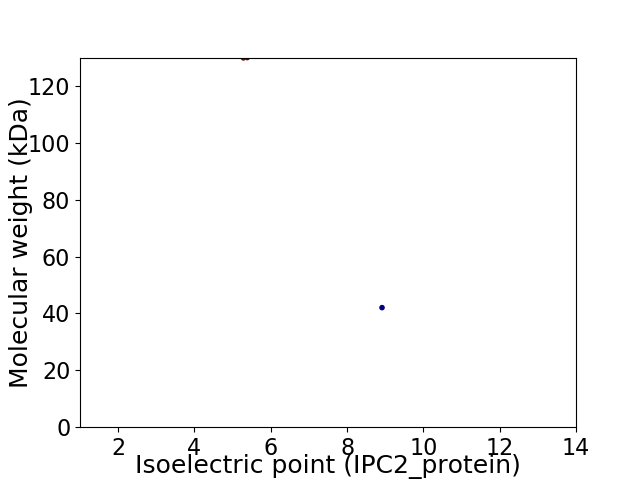
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
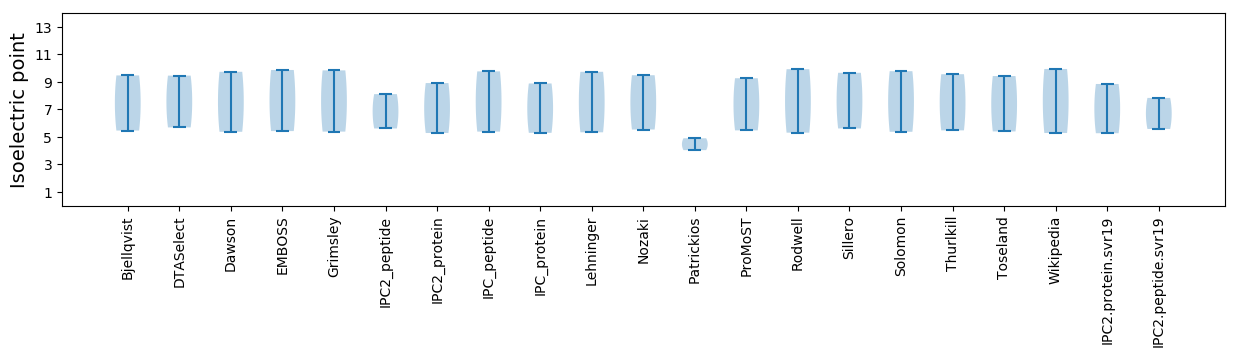
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEC8|A0A1L3KEC8_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 11 OX=1922682 PE=4 SV=1
MM1 pKa = 7.38SVFAQSLGEE10 pKa = 3.88RR11 pKa = 11.84VMEE14 pKa = 4.64LFGTFAGACYY24 pKa = 10.0HH25 pKa = 5.81SARR28 pKa = 11.84KK29 pKa = 8.38FAEE32 pKa = 4.11AADD35 pKa = 3.91YY36 pKa = 10.12QLPSLASIIGYY47 pKa = 9.62VLMGLTFCVAQSLWSWMSPSATKK70 pKa = 9.87TFKK73 pKa = 10.75QFLGACKK80 pKa = 10.22ALWKK84 pKa = 10.45YY85 pKa = 10.48RR86 pKa = 11.84ADD88 pKa = 3.93RR89 pKa = 11.84MQKK92 pKa = 9.94PITGTVVGPVHH103 pKa = 7.41GYY105 pKa = 6.84FTHH108 pKa = 6.74GAEE111 pKa = 3.84HH112 pKa = 7.29YY113 pKa = 8.38YY114 pKa = 10.67RR115 pKa = 11.84CVKK118 pKa = 9.55TSKK121 pKa = 10.61LYY123 pKa = 10.08IVSPGRR129 pKa = 11.84GRR131 pKa = 11.84FYY133 pKa = 10.29IDD135 pKa = 3.05KK136 pKa = 10.62NGEE139 pKa = 3.88EE140 pKa = 4.52KK141 pKa = 11.32YY142 pKa = 10.05EE143 pKa = 4.01VCDD146 pKa = 3.7YY147 pKa = 11.22VHH149 pKa = 6.99EE150 pKa = 4.58LVQSSGIEE158 pKa = 3.86AEE160 pKa = 4.28ANVHH164 pKa = 5.88NGEE167 pKa = 4.23PALPTPALDD176 pKa = 3.44KK177 pKa = 11.21CVVTLAFKK185 pKa = 10.69DD186 pKa = 3.92SNGDD190 pKa = 3.09IHH192 pKa = 7.26NLGHH196 pKa = 5.24GHH198 pKa = 7.21RR199 pKa = 11.84MGDD202 pKa = 3.54YY203 pKa = 10.53LVTNAHH209 pKa = 6.01VNDD212 pKa = 4.17AGLARR217 pKa = 11.84SEE219 pKa = 4.53DD220 pKa = 3.7GQIVFTKK227 pKa = 10.22DD228 pKa = 2.9NQKK231 pKa = 10.35FYY233 pKa = 10.4PAGEE237 pKa = 4.35FIYY240 pKa = 8.77FTKK243 pKa = 10.51NYY245 pKa = 9.35EE246 pKa = 4.09KK247 pKa = 10.59ATGNDD252 pKa = 2.79LGAYY256 pKa = 9.08KK257 pKa = 10.34ISLGTWAASGVTSFQPSNYY276 pKa = 8.91NPQGEE281 pKa = 4.24GRR283 pKa = 11.84IYY285 pKa = 9.79LTAYY289 pKa = 10.42DD290 pKa = 4.07DD291 pKa = 4.12VRR293 pKa = 11.84GKK295 pKa = 9.36TMMAKK300 pKa = 10.3GYY302 pKa = 9.86LHH304 pKa = 6.58NQTKK308 pKa = 9.81EE309 pKa = 3.75DD310 pKa = 4.12CKK312 pKa = 11.05AGLIPHH318 pKa = 6.25TAITRR323 pKa = 11.84AGYY326 pKa = 10.14SGTPIFLMKK335 pKa = 10.06GQRR338 pKa = 11.84RR339 pKa = 11.84TMVGIHH345 pKa = 6.2CGGRR349 pKa = 11.84IDD351 pKa = 4.58GKK353 pKa = 11.23GNYY356 pKa = 8.79GCSIYY361 pKa = 10.88EE362 pKa = 3.87LFQFRR367 pKa = 11.84RR368 pKa = 11.84QLGLEE373 pKa = 3.94PKK375 pKa = 10.2IPLICEE381 pKa = 4.15AKK383 pKa = 10.56YY384 pKa = 10.49ISPPTKK390 pKa = 9.61DD391 pKa = 3.09RR392 pKa = 11.84QYY394 pKa = 11.81DD395 pKa = 3.55RR396 pKa = 11.84YY397 pKa = 10.75DD398 pKa = 3.25QEE400 pKa = 4.2TPEE403 pKa = 3.9QRR405 pKa = 11.84EE406 pKa = 3.78EE407 pKa = 4.21RR408 pKa = 11.84KK409 pKa = 10.42DD410 pKa = 3.5RR411 pKa = 11.84EE412 pKa = 4.4AMEE415 pKa = 4.27KK416 pKa = 10.37AAEE419 pKa = 3.98EE420 pKa = 4.16EE421 pKa = 4.02IRR423 pKa = 11.84EE424 pKa = 4.15NLHH427 pKa = 6.58GEE429 pKa = 4.09ALVPEE434 pKa = 4.57AALDD438 pKa = 4.0GPTWVEE444 pKa = 3.67LGVNTGFEE452 pKa = 3.84IDD454 pKa = 3.26MDD456 pKa = 4.08EE457 pKa = 4.25THH459 pKa = 6.25SQSVGISDD467 pKa = 4.62LGSVSDD473 pKa = 4.57LGLSEE478 pKa = 6.43DD479 pKa = 4.18EE480 pKa = 4.32VQLAAEE486 pKa = 4.0QLPRR490 pKa = 11.84ANLGLNEE497 pKa = 4.66DD498 pKa = 4.46ALQEE502 pKa = 3.89LHH504 pKa = 6.52QDD506 pKa = 2.86QDD508 pKa = 3.43EE509 pKa = 4.34RR510 pKa = 11.84RR511 pKa = 11.84TAAVEE516 pKa = 3.79RR517 pKa = 11.84LRR519 pKa = 11.84TYY521 pKa = 11.23AEE523 pKa = 3.8QQYY526 pKa = 9.58EE527 pKa = 4.57QIPEE531 pKa = 3.98PAPPQEE537 pKa = 4.09QEE539 pKa = 4.07VMSAADD545 pKa = 4.49KK546 pKa = 11.18YY547 pKa = 11.7DD548 pKa = 5.69LIPDD552 pKa = 5.34PIGEE556 pKa = 4.38PEE558 pKa = 4.14FEE560 pKa = 5.04DD561 pKa = 5.25GEE563 pKa = 4.29IEE565 pKa = 4.0EE566 pKa = 4.72PMVHH570 pKa = 6.57FPASGQPVVLGPQVSRR586 pKa = 11.84EE587 pKa = 3.8EE588 pKa = 4.19KK589 pKa = 10.13EE590 pKa = 3.7RR591 pKa = 11.84GEE593 pKa = 3.96RR594 pKa = 11.84TFASRR599 pKa = 11.84ASMPLWVKK607 pKa = 10.54GFLGVGLLQAANVVSPTGKK626 pKa = 7.89TRR628 pKa = 11.84EE629 pKa = 4.14VTDD632 pKa = 3.79EE633 pKa = 3.93QVKK636 pKa = 10.8DD637 pKa = 3.74LLRR640 pKa = 11.84CRR642 pKa = 11.84PQDD645 pKa = 3.25LAAARR650 pKa = 11.84KK651 pKa = 9.38LDD653 pKa = 3.97PQSLLNSTPYY663 pKa = 10.09EE664 pKa = 4.11VYY666 pKa = 10.22RR667 pKa = 11.84SYY669 pKa = 11.2LGCGDD674 pKa = 4.55VEE676 pKa = 4.17WTPDD680 pKa = 3.23GEE682 pKa = 4.96KK683 pKa = 10.45IKK685 pKa = 10.79DD686 pKa = 3.67DD687 pKa = 3.68QDD689 pKa = 3.3NVLAARR695 pKa = 11.84VGKK698 pKa = 10.39SRR700 pKa = 11.84MFSRR704 pKa = 11.84SSNKK708 pKa = 9.46SHH710 pKa = 7.42LPDD713 pKa = 3.69KK714 pKa = 9.92YY715 pKa = 10.75KK716 pKa = 10.86KK717 pKa = 10.46VVADD721 pKa = 4.8LKK723 pKa = 10.98LGPQQGFKK731 pKa = 10.56DD732 pKa = 4.01YY733 pKa = 11.43VMPPTGPEE741 pKa = 3.64AVLGSLRR748 pKa = 11.84TQLKK752 pKa = 7.73KK753 pKa = 9.48TSVEE757 pKa = 3.79PWPKK761 pKa = 10.57EE762 pKa = 3.45FLEE765 pKa = 4.74RR766 pKa = 11.84DD767 pKa = 3.62GGDD770 pKa = 3.05HH771 pKa = 6.98LRR773 pKa = 11.84NEE775 pKa = 4.38LHH777 pKa = 7.5DD778 pKa = 4.27EE779 pKa = 4.07ASKK782 pKa = 11.48YY783 pKa = 9.27PLAYY787 pKa = 9.57EE788 pKa = 4.21AGFLNVHH795 pKa = 6.41DD796 pKa = 4.79FVVKK800 pKa = 10.29LAKK803 pKa = 10.5EE804 pKa = 3.87FDD806 pKa = 3.92GTKK809 pKa = 10.04SAGWSQLYY817 pKa = 10.02RR818 pKa = 11.84PGNKK822 pKa = 9.49AVWQTEE828 pKa = 3.92EE829 pKa = 4.03GLALASYY836 pKa = 7.7MVRR839 pKa = 11.84CRR841 pKa = 11.84LLLRR845 pKa = 11.84MAWGAEE851 pKa = 3.93AMSKK855 pKa = 9.64MSPEE859 pKa = 3.8QMFQNGLSDD868 pKa = 3.78PKK870 pKa = 9.69VASVKK875 pKa = 10.32VEE877 pKa = 3.67PHH879 pKa = 6.6DD880 pKa = 3.99PEE882 pKa = 4.81KK883 pKa = 10.77AAEE886 pKa = 4.01ARR888 pKa = 11.84WRR890 pKa = 11.84VIWGASMIDD899 pKa = 3.73VLTQGVTCRR908 pKa = 11.84MQDD911 pKa = 2.97KK912 pKa = 10.99LDD914 pKa = 3.55ILSYY918 pKa = 10.85QDD920 pKa = 3.13GTGKK924 pKa = 10.01HH925 pKa = 5.08SQGAGLGHH933 pKa = 6.68HH934 pKa = 7.26PEE936 pKa = 5.37GIQMIGKK943 pKa = 9.25HH944 pKa = 5.27IEE946 pKa = 3.76RR947 pKa = 11.84LQASGGDD954 pKa = 3.99LFDD957 pKa = 5.64ADD959 pKa = 3.65ASGWDD964 pKa = 3.64LSVKK968 pKa = 10.04RR969 pKa = 11.84DD970 pKa = 4.03SILMDD975 pKa = 3.29AEE977 pKa = 3.94RR978 pKa = 11.84RR979 pKa = 11.84IRR981 pKa = 11.84CYY983 pKa = 9.72VGPYY987 pKa = 10.07HH988 pKa = 7.13EE989 pKa = 4.68VFKK992 pKa = 10.98DD993 pKa = 3.6LQLADD998 pKa = 3.85AMTNSAHH1005 pKa = 5.71MLVFGEE1011 pKa = 4.33YY1012 pKa = 10.5LFSIYY1017 pKa = 10.37LAGITASGILPTTAQNSFMRR1037 pKa = 11.84AFIARR1042 pKa = 11.84LVGILATFVAGDD1054 pKa = 3.94DD1055 pKa = 3.64LMGAGRR1061 pKa = 11.84FDD1063 pKa = 3.67PEE1065 pKa = 4.25YY1066 pKa = 10.18EE1067 pKa = 4.01RR1068 pKa = 11.84KK1069 pKa = 9.63FGPRR1073 pKa = 11.84EE1074 pKa = 3.54KK1075 pKa = 10.29RR1076 pKa = 11.84ITYY1079 pKa = 9.48RR1080 pKa = 11.84DD1081 pKa = 3.44PGEE1084 pKa = 4.55PIEE1087 pKa = 4.17FTSFLFSKK1095 pKa = 11.01VGDD1098 pKa = 3.24TWIADD1103 pKa = 3.48FTNLGKK1109 pKa = 10.27SCAKK1113 pKa = 10.27LAFSPKK1119 pKa = 9.98SIQPEE1124 pKa = 3.87QLGGVLYY1131 pKa = 10.55HH1132 pKa = 7.01LRR1134 pKa = 11.84DD1135 pKa = 3.79NPAKK1139 pKa = 10.26ADD1141 pKa = 3.82LFRR1144 pKa = 11.84EE1145 pKa = 3.59ICYY1148 pKa = 10.36RR1149 pKa = 11.84MEE1151 pKa = 3.89WPIDD1155 pKa = 3.58QAVAGPDD1162 pKa = 3.81GDD1164 pKa = 4.51CDD1166 pKa = 3.46
MM1 pKa = 7.38SVFAQSLGEE10 pKa = 3.88RR11 pKa = 11.84VMEE14 pKa = 4.64LFGTFAGACYY24 pKa = 10.0HH25 pKa = 5.81SARR28 pKa = 11.84KK29 pKa = 8.38FAEE32 pKa = 4.11AADD35 pKa = 3.91YY36 pKa = 10.12QLPSLASIIGYY47 pKa = 9.62VLMGLTFCVAQSLWSWMSPSATKK70 pKa = 9.87TFKK73 pKa = 10.75QFLGACKK80 pKa = 10.22ALWKK84 pKa = 10.45YY85 pKa = 10.48RR86 pKa = 11.84ADD88 pKa = 3.93RR89 pKa = 11.84MQKK92 pKa = 9.94PITGTVVGPVHH103 pKa = 7.41GYY105 pKa = 6.84FTHH108 pKa = 6.74GAEE111 pKa = 3.84HH112 pKa = 7.29YY113 pKa = 8.38YY114 pKa = 10.67RR115 pKa = 11.84CVKK118 pKa = 9.55TSKK121 pKa = 10.61LYY123 pKa = 10.08IVSPGRR129 pKa = 11.84GRR131 pKa = 11.84FYY133 pKa = 10.29IDD135 pKa = 3.05KK136 pKa = 10.62NGEE139 pKa = 3.88EE140 pKa = 4.52KK141 pKa = 11.32YY142 pKa = 10.05EE143 pKa = 4.01VCDD146 pKa = 3.7YY147 pKa = 11.22VHH149 pKa = 6.99EE150 pKa = 4.58LVQSSGIEE158 pKa = 3.86AEE160 pKa = 4.28ANVHH164 pKa = 5.88NGEE167 pKa = 4.23PALPTPALDD176 pKa = 3.44KK177 pKa = 11.21CVVTLAFKK185 pKa = 10.69DD186 pKa = 3.92SNGDD190 pKa = 3.09IHH192 pKa = 7.26NLGHH196 pKa = 5.24GHH198 pKa = 7.21RR199 pKa = 11.84MGDD202 pKa = 3.54YY203 pKa = 10.53LVTNAHH209 pKa = 6.01VNDD212 pKa = 4.17AGLARR217 pKa = 11.84SEE219 pKa = 4.53DD220 pKa = 3.7GQIVFTKK227 pKa = 10.22DD228 pKa = 2.9NQKK231 pKa = 10.35FYY233 pKa = 10.4PAGEE237 pKa = 4.35FIYY240 pKa = 8.77FTKK243 pKa = 10.51NYY245 pKa = 9.35EE246 pKa = 4.09KK247 pKa = 10.59ATGNDD252 pKa = 2.79LGAYY256 pKa = 9.08KK257 pKa = 10.34ISLGTWAASGVTSFQPSNYY276 pKa = 8.91NPQGEE281 pKa = 4.24GRR283 pKa = 11.84IYY285 pKa = 9.79LTAYY289 pKa = 10.42DD290 pKa = 4.07DD291 pKa = 4.12VRR293 pKa = 11.84GKK295 pKa = 9.36TMMAKK300 pKa = 10.3GYY302 pKa = 9.86LHH304 pKa = 6.58NQTKK308 pKa = 9.81EE309 pKa = 3.75DD310 pKa = 4.12CKK312 pKa = 11.05AGLIPHH318 pKa = 6.25TAITRR323 pKa = 11.84AGYY326 pKa = 10.14SGTPIFLMKK335 pKa = 10.06GQRR338 pKa = 11.84RR339 pKa = 11.84TMVGIHH345 pKa = 6.2CGGRR349 pKa = 11.84IDD351 pKa = 4.58GKK353 pKa = 11.23GNYY356 pKa = 8.79GCSIYY361 pKa = 10.88EE362 pKa = 3.87LFQFRR367 pKa = 11.84RR368 pKa = 11.84QLGLEE373 pKa = 3.94PKK375 pKa = 10.2IPLICEE381 pKa = 4.15AKK383 pKa = 10.56YY384 pKa = 10.49ISPPTKK390 pKa = 9.61DD391 pKa = 3.09RR392 pKa = 11.84QYY394 pKa = 11.81DD395 pKa = 3.55RR396 pKa = 11.84YY397 pKa = 10.75DD398 pKa = 3.25QEE400 pKa = 4.2TPEE403 pKa = 3.9QRR405 pKa = 11.84EE406 pKa = 3.78EE407 pKa = 4.21RR408 pKa = 11.84KK409 pKa = 10.42DD410 pKa = 3.5RR411 pKa = 11.84EE412 pKa = 4.4AMEE415 pKa = 4.27KK416 pKa = 10.37AAEE419 pKa = 3.98EE420 pKa = 4.16EE421 pKa = 4.02IRR423 pKa = 11.84EE424 pKa = 4.15NLHH427 pKa = 6.58GEE429 pKa = 4.09ALVPEE434 pKa = 4.57AALDD438 pKa = 4.0GPTWVEE444 pKa = 3.67LGVNTGFEE452 pKa = 3.84IDD454 pKa = 3.26MDD456 pKa = 4.08EE457 pKa = 4.25THH459 pKa = 6.25SQSVGISDD467 pKa = 4.62LGSVSDD473 pKa = 4.57LGLSEE478 pKa = 6.43DD479 pKa = 4.18EE480 pKa = 4.32VQLAAEE486 pKa = 4.0QLPRR490 pKa = 11.84ANLGLNEE497 pKa = 4.66DD498 pKa = 4.46ALQEE502 pKa = 3.89LHH504 pKa = 6.52QDD506 pKa = 2.86QDD508 pKa = 3.43EE509 pKa = 4.34RR510 pKa = 11.84RR511 pKa = 11.84TAAVEE516 pKa = 3.79RR517 pKa = 11.84LRR519 pKa = 11.84TYY521 pKa = 11.23AEE523 pKa = 3.8QQYY526 pKa = 9.58EE527 pKa = 4.57QIPEE531 pKa = 3.98PAPPQEE537 pKa = 4.09QEE539 pKa = 4.07VMSAADD545 pKa = 4.49KK546 pKa = 11.18YY547 pKa = 11.7DD548 pKa = 5.69LIPDD552 pKa = 5.34PIGEE556 pKa = 4.38PEE558 pKa = 4.14FEE560 pKa = 5.04DD561 pKa = 5.25GEE563 pKa = 4.29IEE565 pKa = 4.0EE566 pKa = 4.72PMVHH570 pKa = 6.57FPASGQPVVLGPQVSRR586 pKa = 11.84EE587 pKa = 3.8EE588 pKa = 4.19KK589 pKa = 10.13EE590 pKa = 3.7RR591 pKa = 11.84GEE593 pKa = 3.96RR594 pKa = 11.84TFASRR599 pKa = 11.84ASMPLWVKK607 pKa = 10.54GFLGVGLLQAANVVSPTGKK626 pKa = 7.89TRR628 pKa = 11.84EE629 pKa = 4.14VTDD632 pKa = 3.79EE633 pKa = 3.93QVKK636 pKa = 10.8DD637 pKa = 3.74LLRR640 pKa = 11.84CRR642 pKa = 11.84PQDD645 pKa = 3.25LAAARR650 pKa = 11.84KK651 pKa = 9.38LDD653 pKa = 3.97PQSLLNSTPYY663 pKa = 10.09EE664 pKa = 4.11VYY666 pKa = 10.22RR667 pKa = 11.84SYY669 pKa = 11.2LGCGDD674 pKa = 4.55VEE676 pKa = 4.17WTPDD680 pKa = 3.23GEE682 pKa = 4.96KK683 pKa = 10.45IKK685 pKa = 10.79DD686 pKa = 3.67DD687 pKa = 3.68QDD689 pKa = 3.3NVLAARR695 pKa = 11.84VGKK698 pKa = 10.39SRR700 pKa = 11.84MFSRR704 pKa = 11.84SSNKK708 pKa = 9.46SHH710 pKa = 7.42LPDD713 pKa = 3.69KK714 pKa = 9.92YY715 pKa = 10.75KK716 pKa = 10.86KK717 pKa = 10.46VVADD721 pKa = 4.8LKK723 pKa = 10.98LGPQQGFKK731 pKa = 10.56DD732 pKa = 4.01YY733 pKa = 11.43VMPPTGPEE741 pKa = 3.64AVLGSLRR748 pKa = 11.84TQLKK752 pKa = 7.73KK753 pKa = 9.48TSVEE757 pKa = 3.79PWPKK761 pKa = 10.57EE762 pKa = 3.45FLEE765 pKa = 4.74RR766 pKa = 11.84DD767 pKa = 3.62GGDD770 pKa = 3.05HH771 pKa = 6.98LRR773 pKa = 11.84NEE775 pKa = 4.38LHH777 pKa = 7.5DD778 pKa = 4.27EE779 pKa = 4.07ASKK782 pKa = 11.48YY783 pKa = 9.27PLAYY787 pKa = 9.57EE788 pKa = 4.21AGFLNVHH795 pKa = 6.41DD796 pKa = 4.79FVVKK800 pKa = 10.29LAKK803 pKa = 10.5EE804 pKa = 3.87FDD806 pKa = 3.92GTKK809 pKa = 10.04SAGWSQLYY817 pKa = 10.02RR818 pKa = 11.84PGNKK822 pKa = 9.49AVWQTEE828 pKa = 3.92EE829 pKa = 4.03GLALASYY836 pKa = 7.7MVRR839 pKa = 11.84CRR841 pKa = 11.84LLLRR845 pKa = 11.84MAWGAEE851 pKa = 3.93AMSKK855 pKa = 9.64MSPEE859 pKa = 3.8QMFQNGLSDD868 pKa = 3.78PKK870 pKa = 9.69VASVKK875 pKa = 10.32VEE877 pKa = 3.67PHH879 pKa = 6.6DD880 pKa = 3.99PEE882 pKa = 4.81KK883 pKa = 10.77AAEE886 pKa = 4.01ARR888 pKa = 11.84WRR890 pKa = 11.84VIWGASMIDD899 pKa = 3.73VLTQGVTCRR908 pKa = 11.84MQDD911 pKa = 2.97KK912 pKa = 10.99LDD914 pKa = 3.55ILSYY918 pKa = 10.85QDD920 pKa = 3.13GTGKK924 pKa = 10.01HH925 pKa = 5.08SQGAGLGHH933 pKa = 6.68HH934 pKa = 7.26PEE936 pKa = 5.37GIQMIGKK943 pKa = 9.25HH944 pKa = 5.27IEE946 pKa = 3.76RR947 pKa = 11.84LQASGGDD954 pKa = 3.99LFDD957 pKa = 5.64ADD959 pKa = 3.65ASGWDD964 pKa = 3.64LSVKK968 pKa = 10.04RR969 pKa = 11.84DD970 pKa = 4.03SILMDD975 pKa = 3.29AEE977 pKa = 3.94RR978 pKa = 11.84RR979 pKa = 11.84IRR981 pKa = 11.84CYY983 pKa = 9.72VGPYY987 pKa = 10.07HH988 pKa = 7.13EE989 pKa = 4.68VFKK992 pKa = 10.98DD993 pKa = 3.6LQLADD998 pKa = 3.85AMTNSAHH1005 pKa = 5.71MLVFGEE1011 pKa = 4.33YY1012 pKa = 10.5LFSIYY1017 pKa = 10.37LAGITASGILPTTAQNSFMRR1037 pKa = 11.84AFIARR1042 pKa = 11.84LVGILATFVAGDD1054 pKa = 3.94DD1055 pKa = 3.64LMGAGRR1061 pKa = 11.84FDD1063 pKa = 3.67PEE1065 pKa = 4.25YY1066 pKa = 10.18EE1067 pKa = 4.01RR1068 pKa = 11.84KK1069 pKa = 9.63FGPRR1073 pKa = 11.84EE1074 pKa = 3.54KK1075 pKa = 10.29RR1076 pKa = 11.84ITYY1079 pKa = 9.48RR1080 pKa = 11.84DD1081 pKa = 3.44PGEE1084 pKa = 4.55PIEE1087 pKa = 4.17FTSFLFSKK1095 pKa = 11.01VGDD1098 pKa = 3.24TWIADD1103 pKa = 3.48FTNLGKK1109 pKa = 10.27SCAKK1113 pKa = 10.27LAFSPKK1119 pKa = 9.98SIQPEE1124 pKa = 3.87QLGGVLYY1131 pKa = 10.55HH1132 pKa = 7.01LRR1134 pKa = 11.84DD1135 pKa = 3.79NPAKK1139 pKa = 10.26ADD1141 pKa = 3.82LFRR1144 pKa = 11.84EE1145 pKa = 3.59ICYY1148 pKa = 10.36RR1149 pKa = 11.84MEE1151 pKa = 3.89WPIDD1155 pKa = 3.58QAVAGPDD1162 pKa = 3.81GDD1164 pKa = 4.51CDD1166 pKa = 3.46
Molecular weight: 129.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEC8|A0A1L3KEC8_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 11 OX=1922682 PE=4 SV=1
MM1 pKa = 7.7EE2 pKa = 4.35NQFTRR7 pKa = 11.84KK8 pKa = 8.2QQAALQKK15 pKa = 10.32AKK17 pKa = 10.15TPAARR22 pKa = 11.84EE23 pKa = 4.05ALKK26 pKa = 10.69KK27 pKa = 10.74LYY29 pKa = 8.74TAQRR33 pKa = 11.84AGTARR38 pKa = 11.84PKK40 pKa = 10.56AQAKK44 pKa = 7.99PRR46 pKa = 11.84PRR48 pKa = 11.84SHH50 pKa = 7.72AIPNFLDD57 pKa = 3.75PMCPFPMPTLASEE70 pKa = 4.7GRR72 pKa = 11.84ALPHH76 pKa = 6.43TGLVSSDD83 pKa = 3.37FTVDD87 pKa = 3.37TTNTTLLLVANTGHH101 pKa = 6.5SGTVAFSVKK110 pKa = 9.89LDD112 pKa = 3.48ASGGVVAGSIEE123 pKa = 4.86TYY125 pKa = 9.98TIPTLSVSDD134 pKa = 4.15HH135 pKa = 6.51NGGPTTSRR143 pKa = 11.84SMKK146 pKa = 10.26LSVSVVNCTNNYY158 pKa = 8.93KK159 pKa = 10.5RR160 pKa = 11.84GGRR163 pKa = 11.84VTYY166 pKa = 10.26INSSQRR172 pKa = 11.84LPPRR176 pKa = 11.84NLDD179 pKa = 3.18LSTEE183 pKa = 4.16YY184 pKa = 11.2ASIVSGIKK192 pKa = 9.43SSPYY196 pKa = 8.67RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84INGEE203 pKa = 3.87DD204 pKa = 3.28LVKK207 pKa = 10.21PKK209 pKa = 10.56HH210 pKa = 6.36LITFPVDD217 pKa = 2.94NKK219 pKa = 10.29VYY221 pKa = 10.41HH222 pKa = 6.64SYY224 pKa = 11.43DD225 pKa = 3.16GFRR228 pKa = 11.84GTITAGDD235 pKa = 3.99FLSYY239 pKa = 10.44MLTPGDD245 pKa = 3.59NVATKK250 pKa = 10.32DD251 pKa = 3.61AEE253 pKa = 4.04ISPRR257 pKa = 11.84PMSTVAWIFDD267 pKa = 4.07PAPDD271 pKa = 3.44EE272 pKa = 3.95QAYY275 pKa = 10.52SFTVRR280 pKa = 11.84ASFYY284 pKa = 9.93TRR286 pKa = 11.84WPLTSVPGQTMHH298 pKa = 6.83NVPTAPAAVLNAVVDD313 pKa = 3.82KK314 pKa = 11.57AEE316 pKa = 3.95AHH318 pKa = 6.25GSDD321 pKa = 4.38LVEE324 pKa = 4.21LAAGGAAVAMGSRR337 pKa = 11.84ISGVARR343 pKa = 11.84SIGSGVQAVGSGIGQALGRR362 pKa = 11.84AFTSPYY368 pKa = 10.19GEE370 pKa = 3.96AVAAGGGEE378 pKa = 4.19GGLLAGMEE386 pKa = 4.58GLSSLQALQLPLLAA400 pKa = 5.95
MM1 pKa = 7.7EE2 pKa = 4.35NQFTRR7 pKa = 11.84KK8 pKa = 8.2QQAALQKK15 pKa = 10.32AKK17 pKa = 10.15TPAARR22 pKa = 11.84EE23 pKa = 4.05ALKK26 pKa = 10.69KK27 pKa = 10.74LYY29 pKa = 8.74TAQRR33 pKa = 11.84AGTARR38 pKa = 11.84PKK40 pKa = 10.56AQAKK44 pKa = 7.99PRR46 pKa = 11.84PRR48 pKa = 11.84SHH50 pKa = 7.72AIPNFLDD57 pKa = 3.75PMCPFPMPTLASEE70 pKa = 4.7GRR72 pKa = 11.84ALPHH76 pKa = 6.43TGLVSSDD83 pKa = 3.37FTVDD87 pKa = 3.37TTNTTLLLVANTGHH101 pKa = 6.5SGTVAFSVKK110 pKa = 9.89LDD112 pKa = 3.48ASGGVVAGSIEE123 pKa = 4.86TYY125 pKa = 9.98TIPTLSVSDD134 pKa = 4.15HH135 pKa = 6.51NGGPTTSRR143 pKa = 11.84SMKK146 pKa = 10.26LSVSVVNCTNNYY158 pKa = 8.93KK159 pKa = 10.5RR160 pKa = 11.84GGRR163 pKa = 11.84VTYY166 pKa = 10.26INSSQRR172 pKa = 11.84LPPRR176 pKa = 11.84NLDD179 pKa = 3.18LSTEE183 pKa = 4.16YY184 pKa = 11.2ASIVSGIKK192 pKa = 9.43SSPYY196 pKa = 8.67RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84INGEE203 pKa = 3.87DD204 pKa = 3.28LVKK207 pKa = 10.21PKK209 pKa = 10.56HH210 pKa = 6.36LITFPVDD217 pKa = 2.94NKK219 pKa = 10.29VYY221 pKa = 10.41HH222 pKa = 6.64SYY224 pKa = 11.43DD225 pKa = 3.16GFRR228 pKa = 11.84GTITAGDD235 pKa = 3.99FLSYY239 pKa = 10.44MLTPGDD245 pKa = 3.59NVATKK250 pKa = 10.32DD251 pKa = 3.61AEE253 pKa = 4.04ISPRR257 pKa = 11.84PMSTVAWIFDD267 pKa = 4.07PAPDD271 pKa = 3.44EE272 pKa = 3.95QAYY275 pKa = 10.52SFTVRR280 pKa = 11.84ASFYY284 pKa = 9.93TRR286 pKa = 11.84WPLTSVPGQTMHH298 pKa = 6.83NVPTAPAAVLNAVVDD313 pKa = 3.82KK314 pKa = 11.57AEE316 pKa = 3.95AHH318 pKa = 6.25GSDD321 pKa = 4.38LVEE324 pKa = 4.21LAAGGAAVAMGSRR337 pKa = 11.84ISGVARR343 pKa = 11.84SIGSGVQAVGSGIGQALGRR362 pKa = 11.84AFTSPYY368 pKa = 10.19GEE370 pKa = 3.96AVAAGGGEE378 pKa = 4.19GGLLAGMEE386 pKa = 4.58GLSSLQALQLPLLAA400 pKa = 5.95
Molecular weight: 42.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1566 |
400 |
1166 |
783.0 |
85.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.387 ± 1.143 | 1.277 ± 0.376 |
5.875 ± 0.907 | 6.513 ± 1.578 |
3.64 ± 0.309 | 9.004 ± 0.24 |
2.363 ± 0.175 | 3.895 ± 0.191 |
5.3 ± 0.629 | 8.365 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.554 ± 0.147 | 2.874 ± 0.424 |
5.811 ± 0.575 | 4.215 ± 0.466 |
5.556 ± 0.027 | 6.705 ± 1.231 |
5.492 ± 1.334 | 6.258 ± 0.601 |
1.149 ± 0.314 | 3.768 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
