
Paraeggerthella hongkongensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Paraeggerthella
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
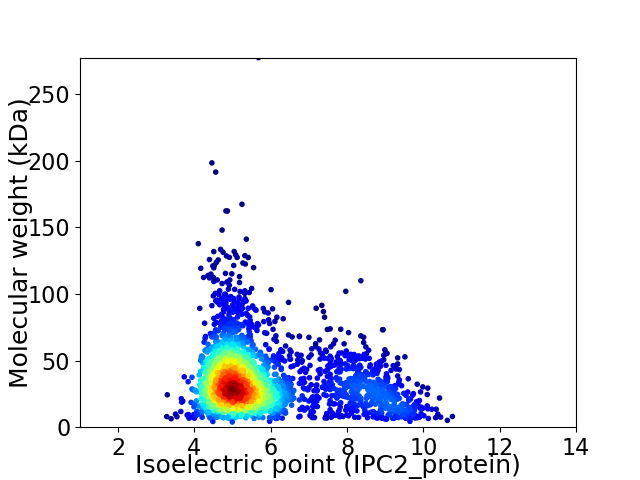
Virtual 2D-PAGE plot for 2391 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369LCR0|A0A369LCR0_9ACTN Uncharacterized protein OS=Paraeggerthella hongkongensis OX=230658 GN=C1879_08450 PE=4 SV=1
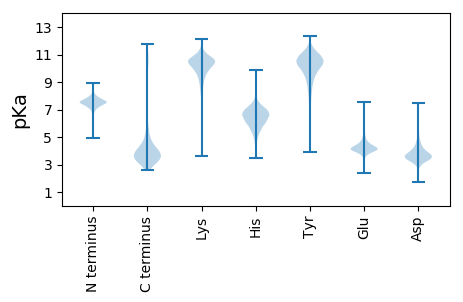
MM1 pKa = 7.48AATVKK6 pKa = 9.88TLLEE10 pKa = 4.14HH11 pKa = 6.97HH12 pKa = 6.94GEE14 pKa = 4.4LYY16 pKa = 10.12WGTRR20 pKa = 11.84QANPGYY26 pKa = 10.35HH27 pKa = 6.35YY28 pKa = 10.98GLSLDD33 pKa = 3.27IGEE36 pKa = 4.91NGVAAQVVYY45 pKa = 11.23VMSDD49 pKa = 2.83LDD51 pKa = 4.13EE52 pKa = 4.76NDD54 pKa = 3.74NPVVTPEE61 pKa = 4.33MLMSCYY67 pKa = 9.42TVADD71 pKa = 4.43LEE73 pKa = 4.68ANGIVPSDD81 pKa = 4.33LMDD84 pKa = 4.17EE85 pKa = 4.36EE86 pKa = 6.48DD87 pKa = 3.65NDD89 pKa = 4.58SVKK92 pKa = 10.37GWYY95 pKa = 9.95CIEE98 pKa = 3.85EE99 pKa = 4.3SFFPEE104 pKa = 4.01NQVEE108 pKa = 4.63ALQEE112 pKa = 4.29SNAQLDD118 pKa = 4.37GYY120 pKa = 10.84LDD122 pKa = 3.43IVLDD126 pKa = 3.59IMLISPEE133 pKa = 3.88EE134 pKa = 4.05VAAGMPSLDD143 pKa = 4.78DD144 pKa = 3.5ISFFDD149 pKa = 5.06LLSEE153 pKa = 4.31LEE155 pKa = 4.84GIAEE159 pKa = 4.78RR160 pKa = 11.84IDD162 pKa = 3.43SGALAKK168 pKa = 9.83PLPFGFRR175 pKa = 11.84LVGDD179 pKa = 4.34ALFGWEE185 pKa = 4.64FADD188 pKa = 4.53EE189 pKa = 4.2EE190 pKa = 4.4DD191 pKa = 3.51WRR193 pKa = 5.44
MM1 pKa = 7.48AATVKK6 pKa = 9.88TLLEE10 pKa = 4.14HH11 pKa = 6.97HH12 pKa = 6.94GEE14 pKa = 4.4LYY16 pKa = 10.12WGTRR20 pKa = 11.84QANPGYY26 pKa = 10.35HH27 pKa = 6.35YY28 pKa = 10.98GLSLDD33 pKa = 3.27IGEE36 pKa = 4.91NGVAAQVVYY45 pKa = 11.23VMSDD49 pKa = 2.83LDD51 pKa = 4.13EE52 pKa = 4.76NDD54 pKa = 3.74NPVVTPEE61 pKa = 4.33MLMSCYY67 pKa = 9.42TVADD71 pKa = 4.43LEE73 pKa = 4.68ANGIVPSDD81 pKa = 4.33LMDD84 pKa = 4.17EE85 pKa = 4.36EE86 pKa = 6.48DD87 pKa = 3.65NDD89 pKa = 4.58SVKK92 pKa = 10.37GWYY95 pKa = 9.95CIEE98 pKa = 3.85EE99 pKa = 4.3SFFPEE104 pKa = 4.01NQVEE108 pKa = 4.63ALQEE112 pKa = 4.29SNAQLDD118 pKa = 4.37GYY120 pKa = 10.84LDD122 pKa = 3.43IVLDD126 pKa = 3.59IMLISPEE133 pKa = 3.88EE134 pKa = 4.05VAAGMPSLDD143 pKa = 4.78DD144 pKa = 3.5ISFFDD149 pKa = 5.06LLSEE153 pKa = 4.31LEE155 pKa = 4.84GIAEE159 pKa = 4.78RR160 pKa = 11.84IDD162 pKa = 3.43SGALAKK168 pKa = 9.83PLPFGFRR175 pKa = 11.84LVGDD179 pKa = 4.34ALFGWEE185 pKa = 4.64FADD188 pKa = 4.53EE189 pKa = 4.2EE190 pKa = 4.4DD191 pKa = 3.51WRR193 pKa = 5.44
Molecular weight: 21.39 kDa
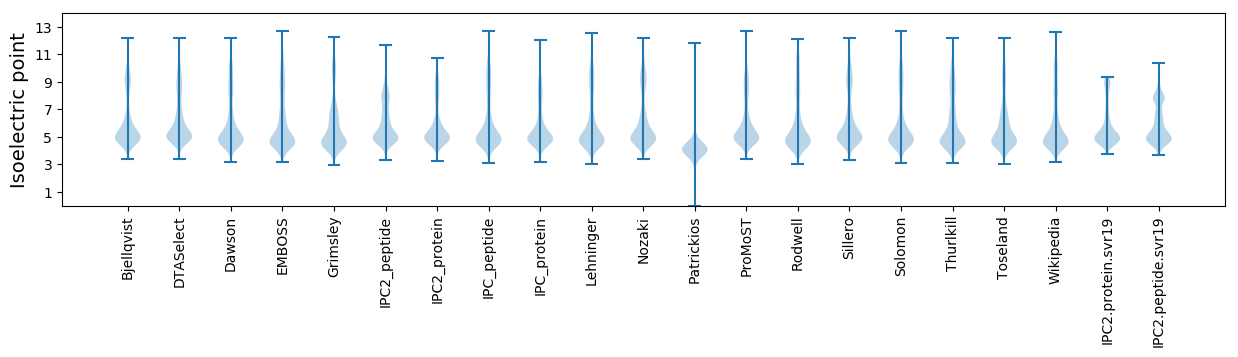
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369L4U7|A0A369L4U7_9ACTN FHA domain-containing protein OS=Paraeggerthella hongkongensis OX=230658 GN=C1879_11905 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84AARR5 pKa = 11.84CASRR9 pKa = 11.84RR10 pKa = 11.84NARR13 pKa = 11.84RR14 pKa = 11.84ANALLLALGILLIASALASLMLGRR38 pKa = 11.84YY39 pKa = 8.03PITPVEE45 pKa = 4.08AVGMLANLAFPVDD58 pKa = 4.35PFWTPQQEE66 pKa = 4.42TLFFQVRR73 pKa = 11.84LPRR76 pKa = 11.84IVLAVMVGCSLATAGAAYY94 pKa = 10.13QGTFQNPLVSPDD106 pKa = 3.37ILGASQGAAFGAACAILLGMGAFGVSASAFLFSIAAVMLVLLIGTRR152 pKa = 11.84ARR154 pKa = 11.84GNHH157 pKa = 5.8LLIVVLAGVMVSSLFSAGVSFTKK180 pKa = 10.9LIADD184 pKa = 4.55PNNQLAAITYY194 pKa = 8.41WLMGSLTGAKK204 pKa = 9.05WSDD207 pKa = 3.22MALAAIPMAVGLGALFALRR226 pKa = 11.84WRR228 pKa = 11.84INVLTMGDD236 pKa = 4.25DD237 pKa = 3.74EE238 pKa = 5.72ASTMGVNARR247 pKa = 11.84RR248 pKa = 11.84TRR250 pKa = 11.84LIVILAATLITASSVAVSGMIGWVGLVIPHH280 pKa = 7.26LSRR283 pKa = 11.84MVIGCDD289 pKa = 3.48YY290 pKa = 11.05RR291 pKa = 11.84KK292 pKa = 10.11LLPASMLMGASFMLIVDD309 pKa = 4.85DD310 pKa = 4.32IARR313 pKa = 11.84LVATAEE319 pKa = 3.94IPIGILTAFVGAPFFLYY336 pKa = 10.89LITRR340 pKa = 11.84KK341 pKa = 9.39KK342 pKa = 10.44QKK344 pKa = 10.45LL345 pKa = 3.45
MM1 pKa = 7.62RR2 pKa = 11.84AARR5 pKa = 11.84CASRR9 pKa = 11.84RR10 pKa = 11.84NARR13 pKa = 11.84RR14 pKa = 11.84ANALLLALGILLIASALASLMLGRR38 pKa = 11.84YY39 pKa = 8.03PITPVEE45 pKa = 4.08AVGMLANLAFPVDD58 pKa = 4.35PFWTPQQEE66 pKa = 4.42TLFFQVRR73 pKa = 11.84LPRR76 pKa = 11.84IVLAVMVGCSLATAGAAYY94 pKa = 10.13QGTFQNPLVSPDD106 pKa = 3.37ILGASQGAAFGAACAILLGMGAFGVSASAFLFSIAAVMLVLLIGTRR152 pKa = 11.84ARR154 pKa = 11.84GNHH157 pKa = 5.8LLIVVLAGVMVSSLFSAGVSFTKK180 pKa = 10.9LIADD184 pKa = 4.55PNNQLAAITYY194 pKa = 8.41WLMGSLTGAKK204 pKa = 9.05WSDD207 pKa = 3.22MALAAIPMAVGLGALFALRR226 pKa = 11.84WRR228 pKa = 11.84INVLTMGDD236 pKa = 4.25DD237 pKa = 3.74EE238 pKa = 5.72ASTMGVNARR247 pKa = 11.84RR248 pKa = 11.84TRR250 pKa = 11.84LIVILAATLITASSVAVSGMIGWVGLVIPHH280 pKa = 7.26LSRR283 pKa = 11.84MVIGCDD289 pKa = 3.48YY290 pKa = 11.05RR291 pKa = 11.84KK292 pKa = 10.11LLPASMLMGASFMLIVDD309 pKa = 4.85DD310 pKa = 4.32IARR313 pKa = 11.84LVATAEE319 pKa = 3.94IPIGILTAFVGAPFFLYY336 pKa = 10.89LITRR340 pKa = 11.84KK341 pKa = 9.39KK342 pKa = 10.44QKK344 pKa = 10.45LL345 pKa = 3.45
Molecular weight: 36.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
817088 |
37 |
2536 |
341.7 |
37.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.976 ± 0.07 | 1.593 ± 0.023 |
6.083 ± 0.039 | 6.303 ± 0.047 |
3.923 ± 0.035 | 8.18 ± 0.052 |
1.73 ± 0.018 | 4.811 ± 0.048 |
3.806 ± 0.041 | 9.381 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.671 ± 0.026 | 2.84 ± 0.028 |
4.309 ± 0.029 | 2.955 ± 0.026 |
6.02 ± 0.056 | 6.096 ± 0.045 |
5.061 ± 0.032 | 8.355 ± 0.047 |
1.08 ± 0.021 | 2.827 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
