
Kern Canyon virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Kern Canyon ledantevirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
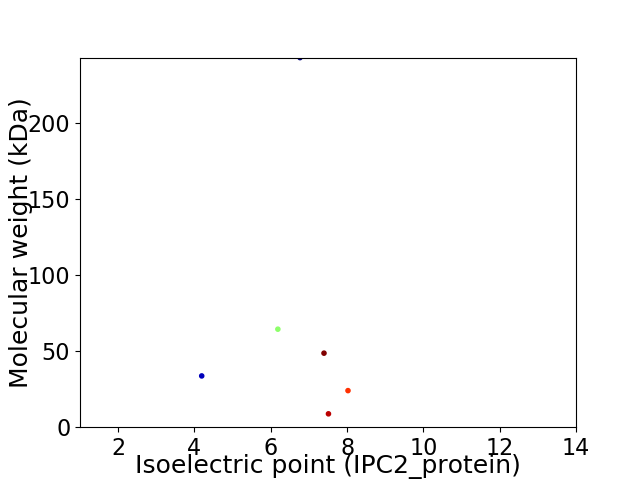
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
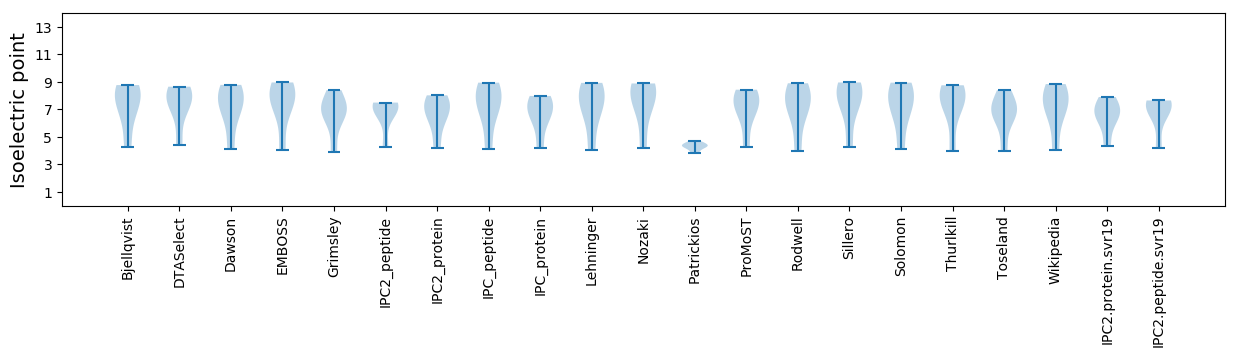
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R1K7|A0A0D3R1K7_9RHAB Phosphoprotein OS=Kern Canyon virus OX=380433 PE=4 SV=1
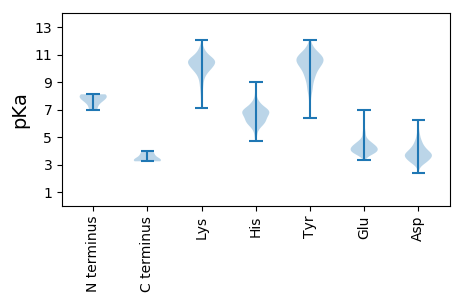
MM1 pKa = 7.76SDD3 pKa = 3.81FRR5 pKa = 11.84DD6 pKa = 3.78KK7 pKa = 11.19VKK9 pKa = 10.82KK10 pKa = 10.23LAAKK14 pKa = 9.64DD15 pKa = 3.08IWDD18 pKa = 4.73SISKK22 pKa = 10.83LEE24 pKa = 3.88IDD26 pKa = 4.17EE27 pKa = 5.56DD28 pKa = 4.09EE29 pKa = 4.54EE30 pKa = 4.96STFLEE35 pKa = 4.44NEE37 pKa = 4.05EE38 pKa = 4.9KK39 pKa = 10.45PRR41 pKa = 11.84SDD43 pKa = 5.12DD44 pKa = 3.02NWTKK48 pKa = 11.03AEE50 pKa = 4.26EE51 pKa = 4.22PSEE54 pKa = 4.29VPAYY58 pKa = 10.04EE59 pKa = 4.31EE60 pKa = 4.16EE61 pKa = 4.62FDD63 pKa = 4.66TEE65 pKa = 5.33DD66 pKa = 3.6EE67 pKa = 4.48DD68 pKa = 5.2EE69 pKa = 5.52EE70 pKa = 4.14IAQACKK76 pKa = 10.56EE77 pKa = 4.31KK78 pKa = 10.96GPEE81 pKa = 3.91DD82 pKa = 3.86LLEE85 pKa = 4.66GGILDD90 pKa = 3.76LHH92 pKa = 6.9AGSEE96 pKa = 4.32FEE98 pKa = 6.5DD99 pKa = 5.66DD100 pKa = 3.85DD101 pKa = 4.27THH103 pKa = 8.47CLGLSDD109 pKa = 4.83PLEE112 pKa = 4.15LSIDD116 pKa = 3.76SQEE119 pKa = 4.14RR120 pKa = 11.84AQSSEE125 pKa = 3.68DD126 pKa = 3.69DD127 pKa = 3.93NNEE130 pKa = 3.85GVPMVLPRR138 pKa = 11.84ILLDD142 pKa = 3.52TVGSQEE148 pKa = 4.9DD149 pKa = 3.86LDD151 pKa = 4.21DD152 pKa = 3.66RR153 pKa = 11.84CYY155 pKa = 11.76AMLHH159 pKa = 5.35MAFKK163 pKa = 10.84KK164 pKa = 10.21LGWMLLPQTCHH175 pKa = 6.87PIDD178 pKa = 3.82EE179 pKa = 4.71ALHH182 pKa = 6.11LVSTPIRR189 pKa = 11.84VQRR192 pKa = 11.84SQISSVSDD200 pKa = 3.38NPPSLEE206 pKa = 3.76NSTSVNQGRR215 pKa = 11.84SSEE218 pKa = 4.32AAPQSNSAVRR228 pKa = 11.84FEE230 pKa = 4.88DD231 pKa = 3.55IIRR234 pKa = 11.84RR235 pKa = 11.84MEE237 pKa = 3.98TGDD240 pKa = 3.08IKK242 pKa = 10.93FPKK245 pKa = 10.05KK246 pKa = 10.1NGKK249 pKa = 10.39GYY251 pKa = 7.52MTLDD255 pKa = 3.52LGIPGLTPEE264 pKa = 5.8LIRR267 pKa = 11.84EE268 pKa = 4.45VIAITSDD275 pKa = 3.26EE276 pKa = 4.38ADD278 pKa = 5.05LISKK282 pKa = 10.26LLDD285 pKa = 3.29ILDD288 pKa = 3.63MKK290 pKa = 10.27EE291 pKa = 4.31YY292 pKa = 10.73IMTLCHH298 pKa = 6.34YY299 pKa = 8.95PP300 pKa = 3.25
MM1 pKa = 7.76SDD3 pKa = 3.81FRR5 pKa = 11.84DD6 pKa = 3.78KK7 pKa = 11.19VKK9 pKa = 10.82KK10 pKa = 10.23LAAKK14 pKa = 9.64DD15 pKa = 3.08IWDD18 pKa = 4.73SISKK22 pKa = 10.83LEE24 pKa = 3.88IDD26 pKa = 4.17EE27 pKa = 5.56DD28 pKa = 4.09EE29 pKa = 4.54EE30 pKa = 4.96STFLEE35 pKa = 4.44NEE37 pKa = 4.05EE38 pKa = 4.9KK39 pKa = 10.45PRR41 pKa = 11.84SDD43 pKa = 5.12DD44 pKa = 3.02NWTKK48 pKa = 11.03AEE50 pKa = 4.26EE51 pKa = 4.22PSEE54 pKa = 4.29VPAYY58 pKa = 10.04EE59 pKa = 4.31EE60 pKa = 4.16EE61 pKa = 4.62FDD63 pKa = 4.66TEE65 pKa = 5.33DD66 pKa = 3.6EE67 pKa = 4.48DD68 pKa = 5.2EE69 pKa = 5.52EE70 pKa = 4.14IAQACKK76 pKa = 10.56EE77 pKa = 4.31KK78 pKa = 10.96GPEE81 pKa = 3.91DD82 pKa = 3.86LLEE85 pKa = 4.66GGILDD90 pKa = 3.76LHH92 pKa = 6.9AGSEE96 pKa = 4.32FEE98 pKa = 6.5DD99 pKa = 5.66DD100 pKa = 3.85DD101 pKa = 4.27THH103 pKa = 8.47CLGLSDD109 pKa = 4.83PLEE112 pKa = 4.15LSIDD116 pKa = 3.76SQEE119 pKa = 4.14RR120 pKa = 11.84AQSSEE125 pKa = 3.68DD126 pKa = 3.69DD127 pKa = 3.93NNEE130 pKa = 3.85GVPMVLPRR138 pKa = 11.84ILLDD142 pKa = 3.52TVGSQEE148 pKa = 4.9DD149 pKa = 3.86LDD151 pKa = 4.21DD152 pKa = 3.66RR153 pKa = 11.84CYY155 pKa = 11.76AMLHH159 pKa = 5.35MAFKK163 pKa = 10.84KK164 pKa = 10.21LGWMLLPQTCHH175 pKa = 6.87PIDD178 pKa = 3.82EE179 pKa = 4.71ALHH182 pKa = 6.11LVSTPIRR189 pKa = 11.84VQRR192 pKa = 11.84SQISSVSDD200 pKa = 3.38NPPSLEE206 pKa = 3.76NSTSVNQGRR215 pKa = 11.84SSEE218 pKa = 4.32AAPQSNSAVRR228 pKa = 11.84FEE230 pKa = 4.88DD231 pKa = 3.55IIRR234 pKa = 11.84RR235 pKa = 11.84MEE237 pKa = 3.98TGDD240 pKa = 3.08IKK242 pKa = 10.93FPKK245 pKa = 10.05KK246 pKa = 10.1NGKK249 pKa = 10.39GYY251 pKa = 7.52MTLDD255 pKa = 3.52LGIPGLTPEE264 pKa = 5.8LIRR267 pKa = 11.84EE268 pKa = 4.45VIAITSDD275 pKa = 3.26EE276 pKa = 4.38ADD278 pKa = 5.05LISKK282 pKa = 10.26LLDD285 pKa = 3.29ILDD288 pKa = 3.63MKK290 pKa = 10.27EE291 pKa = 4.31YY292 pKa = 10.73IMTLCHH298 pKa = 6.34YY299 pKa = 8.95PP300 pKa = 3.25
Molecular weight: 33.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R1G9|A0A0D3R1G9_9RHAB GDP polyribonucleotidyltransferase OS=Kern Canyon virus OX=380433 PE=4 SV=1
MM1 pKa = 7.58LSKK4 pKa = 10.63LKK6 pKa = 10.7KK7 pKa = 10.02SFSKK11 pKa = 10.77KK12 pKa = 8.85EE13 pKa = 3.88SEE15 pKa = 4.1KK16 pKa = 10.41TVMIPEE22 pKa = 5.18GPPPDD27 pKa = 3.69YY28 pKa = 11.1YY29 pKa = 11.53GGFFYY34 pKa = 10.23PEE36 pKa = 3.96PSAPPDD42 pKa = 3.38EE43 pKa = 4.35LVKK46 pKa = 10.47TEE48 pKa = 3.96SLYY51 pKa = 11.06VKK53 pKa = 10.78AEE55 pKa = 3.85IVVKK59 pKa = 10.68SEE61 pKa = 3.99VPIKK65 pKa = 10.32EE66 pKa = 3.91MKK68 pKa = 10.35ALQEE72 pKa = 4.02ILAVWVDD79 pKa = 3.8KK80 pKa = 11.2NVSPVQQKK88 pKa = 10.33HH89 pKa = 5.84LDD91 pKa = 3.08TWFYY95 pKa = 11.34LCLGLHH101 pKa = 5.62VRR103 pKa = 11.84RR104 pKa = 11.84DD105 pKa = 3.51QDD107 pKa = 3.44CTYY110 pKa = 10.47TNMYY114 pKa = 9.04RR115 pKa = 11.84AAVDD119 pKa = 3.26MVVEE123 pKa = 4.58VNHH126 pKa = 6.4RR127 pKa = 11.84PKK129 pKa = 10.14TSSAIKK135 pKa = 9.58YY136 pKa = 9.97IPFEE140 pKa = 3.99QTFDD144 pKa = 3.59TIYY147 pKa = 10.77GGRR150 pKa = 11.84PCEE153 pKa = 3.66VSFKK157 pKa = 11.3NKK159 pKa = 8.96MVSSKK164 pKa = 10.84RR165 pKa = 11.84KK166 pKa = 7.36GTPAHH171 pKa = 6.82LLYY174 pKa = 10.71NQPLKK179 pKa = 10.92NGQSLPKK186 pKa = 9.79PVEE189 pKa = 3.64IFNGYY194 pKa = 8.44GVEE197 pKa = 5.24VKK199 pKa = 9.85LTDD202 pKa = 3.67NKK204 pKa = 10.0VHH206 pKa = 6.8EE207 pKa = 4.67ISLII211 pKa = 3.73
MM1 pKa = 7.58LSKK4 pKa = 10.63LKK6 pKa = 10.7KK7 pKa = 10.02SFSKK11 pKa = 10.77KK12 pKa = 8.85EE13 pKa = 3.88SEE15 pKa = 4.1KK16 pKa = 10.41TVMIPEE22 pKa = 5.18GPPPDD27 pKa = 3.69YY28 pKa = 11.1YY29 pKa = 11.53GGFFYY34 pKa = 10.23PEE36 pKa = 3.96PSAPPDD42 pKa = 3.38EE43 pKa = 4.35LVKK46 pKa = 10.47TEE48 pKa = 3.96SLYY51 pKa = 11.06VKK53 pKa = 10.78AEE55 pKa = 3.85IVVKK59 pKa = 10.68SEE61 pKa = 3.99VPIKK65 pKa = 10.32EE66 pKa = 3.91MKK68 pKa = 10.35ALQEE72 pKa = 4.02ILAVWVDD79 pKa = 3.8KK80 pKa = 11.2NVSPVQQKK88 pKa = 10.33HH89 pKa = 5.84LDD91 pKa = 3.08TWFYY95 pKa = 11.34LCLGLHH101 pKa = 5.62VRR103 pKa = 11.84RR104 pKa = 11.84DD105 pKa = 3.51QDD107 pKa = 3.44CTYY110 pKa = 10.47TNMYY114 pKa = 9.04RR115 pKa = 11.84AAVDD119 pKa = 3.26MVVEE123 pKa = 4.58VNHH126 pKa = 6.4RR127 pKa = 11.84PKK129 pKa = 10.14TSSAIKK135 pKa = 9.58YY136 pKa = 9.97IPFEE140 pKa = 3.99QTFDD144 pKa = 3.59TIYY147 pKa = 10.77GGRR150 pKa = 11.84PCEE153 pKa = 3.66VSFKK157 pKa = 11.3NKK159 pKa = 8.96MVSSKK164 pKa = 10.84RR165 pKa = 11.84KK166 pKa = 7.36GTPAHH171 pKa = 6.82LLYY174 pKa = 10.71NQPLKK179 pKa = 10.92NGQSLPKK186 pKa = 9.79PVEE189 pKa = 3.64IFNGYY194 pKa = 8.44GVEE197 pKa = 5.24VKK199 pKa = 9.85LTDD202 pKa = 3.67NKK204 pKa = 10.0VHH206 pKa = 6.8EE207 pKa = 4.67ISLII211 pKa = 3.73
Molecular weight: 24.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3706 |
77 |
2129 |
617.7 |
70.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.263 ± 0.577 | 1.781 ± 0.279 |
6.557 ± 0.581 | 6.746 ± 0.974 |
4.452 ± 0.633 | 5.828 ± 0.365 |
2.563 ± 0.392 | 7.528 ± 0.576 |
7.501 ± 0.724 | 9.552 ± 0.817 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.386 | 4.155 ± 0.332 |
5.046 ± 0.489 | 2.914 ± 0.461 |
4.857 ± 0.48 | 7.879 ± 0.498 |
5.181 ± 0.352 | 5.505 ± 0.759 |
1.727 ± 0.212 | 3.427 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
