
Anaeromassilibacillus sp. An172
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Anaeromassilibacillus; unclassified Anaeromassilibacillus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
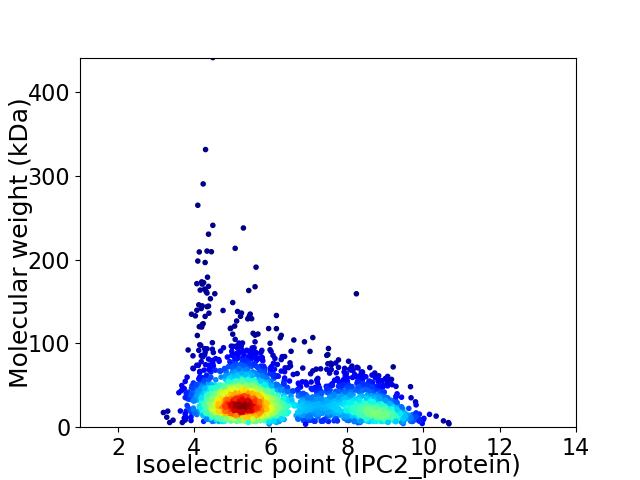
Virtual 2D-PAGE plot for 2508 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4N5S7|A0A1Y4N5S7_9FIRM Conjugal transfer protein OS=Anaeromassilibacillus sp. An172 OX=1965570 GN=B5F08_10600 PE=4 SV=1
MM1 pKa = 7.23FVLNDD6 pKa = 3.53YY7 pKa = 8.2MKK9 pKa = 10.91KK10 pKa = 9.57NLRR13 pKa = 11.84FIIPVAALIVLAIIFGIMYY32 pKa = 10.12IVNNNSDD39 pKa = 3.64QDD41 pKa = 3.89NTDD44 pKa = 2.84TTEE47 pKa = 4.16PVSTEE52 pKa = 3.71PQTTMTATEE61 pKa = 4.36EE62 pKa = 3.82LSQYY66 pKa = 10.6KK67 pKa = 9.9IGIMQFDD74 pKa = 4.23DD75 pKa = 3.62SQEE78 pKa = 3.98YY79 pKa = 10.69SDD81 pKa = 4.99VYY83 pKa = 11.53DD84 pKa = 3.86EE85 pKa = 5.23FVLAMEE91 pKa = 4.65HH92 pKa = 6.82RR93 pKa = 11.84GFTEE97 pKa = 4.19GVNVEE102 pKa = 4.23YY103 pKa = 11.05VYY105 pKa = 10.96MNAEE109 pKa = 4.18GDD111 pKa = 4.0SEE113 pKa = 4.46KK114 pKa = 10.86CSDD117 pKa = 3.54MAQSIVDD124 pKa = 3.92SGVDD128 pKa = 3.82LIFAISEE135 pKa = 4.16EE136 pKa = 4.29NAVAAYY142 pKa = 9.25NATKK146 pKa = 10.22DD147 pKa = 3.3IPIVFGAVNDD157 pKa = 4.13PEE159 pKa = 4.18EE160 pKa = 5.83AEE162 pKa = 5.39LIDD165 pKa = 3.85TNEE168 pKa = 3.8APGANVTGVSDD179 pKa = 3.86YY180 pKa = 11.27SPSIEE185 pKa = 4.56QIDD188 pKa = 4.38LIKK191 pKa = 10.15TVFPEE196 pKa = 4.1AKK198 pKa = 9.19TVGAVYY204 pKa = 10.8DD205 pKa = 4.08EE206 pKa = 4.98LNEE209 pKa = 4.18SSVTQISLAEE219 pKa = 4.14TQAQTNEE226 pKa = 4.02MTFDD230 pKa = 3.85KK231 pKa = 11.17YY232 pKa = 10.83PVNDD236 pKa = 3.56TTDD239 pKa = 3.7FDD241 pKa = 5.13SYY243 pKa = 9.4ITDD246 pKa = 3.79MIEE249 pKa = 4.07KK250 pKa = 9.88VDD252 pKa = 4.55VIYY255 pKa = 10.87LPYY258 pKa = 10.69DD259 pKa = 3.32EE260 pKa = 6.01LLIDD264 pKa = 4.96NIDD267 pKa = 4.16KK268 pKa = 9.18ITAVAYY274 pKa = 7.87EE275 pKa = 3.97HH276 pKa = 7.1GIPVVGGNEE285 pKa = 3.68EE286 pKa = 4.3MVQKK290 pKa = 10.75GCTIAYY296 pKa = 9.26GIDD299 pKa = 3.45YY300 pKa = 10.75ADD302 pKa = 3.73VGKK305 pKa = 10.55QSALMAVEE313 pKa = 4.65ILQNDD318 pKa = 4.23GTPAEE323 pKa = 4.23MPVRR327 pKa = 11.84YY328 pKa = 9.73LNEE331 pKa = 4.67LILYY335 pKa = 9.27VNTEE339 pKa = 3.95AKK341 pKa = 9.83EE342 pKa = 3.95KK343 pKa = 10.96LGFEE347 pKa = 5.02LDD349 pKa = 3.12EE350 pKa = 4.28TLQNKK355 pKa = 9.13AVLLPEE361 pKa = 4.35KK362 pKa = 10.7SEE364 pKa = 4.08EE365 pKa = 3.98
MM1 pKa = 7.23FVLNDD6 pKa = 3.53YY7 pKa = 8.2MKK9 pKa = 10.91KK10 pKa = 9.57NLRR13 pKa = 11.84FIIPVAALIVLAIIFGIMYY32 pKa = 10.12IVNNNSDD39 pKa = 3.64QDD41 pKa = 3.89NTDD44 pKa = 2.84TTEE47 pKa = 4.16PVSTEE52 pKa = 3.71PQTTMTATEE61 pKa = 4.36EE62 pKa = 3.82LSQYY66 pKa = 10.6KK67 pKa = 9.9IGIMQFDD74 pKa = 4.23DD75 pKa = 3.62SQEE78 pKa = 3.98YY79 pKa = 10.69SDD81 pKa = 4.99VYY83 pKa = 11.53DD84 pKa = 3.86EE85 pKa = 5.23FVLAMEE91 pKa = 4.65HH92 pKa = 6.82RR93 pKa = 11.84GFTEE97 pKa = 4.19GVNVEE102 pKa = 4.23YY103 pKa = 11.05VYY105 pKa = 10.96MNAEE109 pKa = 4.18GDD111 pKa = 4.0SEE113 pKa = 4.46KK114 pKa = 10.86CSDD117 pKa = 3.54MAQSIVDD124 pKa = 3.92SGVDD128 pKa = 3.82LIFAISEE135 pKa = 4.16EE136 pKa = 4.29NAVAAYY142 pKa = 9.25NATKK146 pKa = 10.22DD147 pKa = 3.3IPIVFGAVNDD157 pKa = 4.13PEE159 pKa = 4.18EE160 pKa = 5.83AEE162 pKa = 5.39LIDD165 pKa = 3.85TNEE168 pKa = 3.8APGANVTGVSDD179 pKa = 3.86YY180 pKa = 11.27SPSIEE185 pKa = 4.56QIDD188 pKa = 4.38LIKK191 pKa = 10.15TVFPEE196 pKa = 4.1AKK198 pKa = 9.19TVGAVYY204 pKa = 10.8DD205 pKa = 4.08EE206 pKa = 4.98LNEE209 pKa = 4.18SSVTQISLAEE219 pKa = 4.14TQAQTNEE226 pKa = 4.02MTFDD230 pKa = 3.85KK231 pKa = 11.17YY232 pKa = 10.83PVNDD236 pKa = 3.56TTDD239 pKa = 3.7FDD241 pKa = 5.13SYY243 pKa = 9.4ITDD246 pKa = 3.79MIEE249 pKa = 4.07KK250 pKa = 9.88VDD252 pKa = 4.55VIYY255 pKa = 10.87LPYY258 pKa = 10.69DD259 pKa = 3.32EE260 pKa = 6.01LLIDD264 pKa = 4.96NIDD267 pKa = 4.16KK268 pKa = 9.18ITAVAYY274 pKa = 7.87EE275 pKa = 3.97HH276 pKa = 7.1GIPVVGGNEE285 pKa = 3.68EE286 pKa = 4.3MVQKK290 pKa = 10.75GCTIAYY296 pKa = 9.26GIDD299 pKa = 3.45YY300 pKa = 10.75ADD302 pKa = 3.73VGKK305 pKa = 10.55QSALMAVEE313 pKa = 4.65ILQNDD318 pKa = 4.23GTPAEE323 pKa = 4.23MPVRR327 pKa = 11.84YY328 pKa = 9.73LNEE331 pKa = 4.67LILYY335 pKa = 9.27VNTEE339 pKa = 3.95AKK341 pKa = 9.83EE342 pKa = 3.95KK343 pKa = 10.96LGFEE347 pKa = 5.02LDD349 pKa = 3.12EE350 pKa = 4.28TLQNKK355 pKa = 9.13AVLLPEE361 pKa = 4.35KK362 pKa = 10.7SEE364 pKa = 4.08EE365 pKa = 3.98
Molecular weight: 40.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4NDK2|A0A1Y4NDK2_9FIRM Pseudouridine synthase OS=Anaeromassilibacillus sp. An172 OX=1965570 GN=B5F08_05790 PE=4 SV=1
MM1 pKa = 7.9AEE3 pKa = 3.97RR4 pKa = 11.84PMNRR8 pKa = 11.84SRR10 pKa = 11.84KK11 pKa = 8.43PRR13 pKa = 11.84KK14 pKa = 8.99KK15 pKa = 10.03VCGFCVDD22 pKa = 3.21KK23 pKa = 11.52VEE25 pKa = 4.75NIDD28 pKa = 3.57YY29 pKa = 10.94KK30 pKa = 11.15DD31 pKa = 3.08IARR34 pKa = 11.84LRR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 8.42MSEE41 pKa = 3.7RR42 pKa = 11.84GKK44 pKa = 10.14ILPRR48 pKa = 11.84RR49 pKa = 11.84VTGTCAAHH57 pKa = 5.98QRR59 pKa = 11.84ALTVAIKK66 pKa = 10.31RR67 pKa = 11.84ARR69 pKa = 11.84HH70 pKa = 5.37LALLPYY76 pKa = 8.42TTDD79 pKa = 2.85
MM1 pKa = 7.9AEE3 pKa = 3.97RR4 pKa = 11.84PMNRR8 pKa = 11.84SRR10 pKa = 11.84KK11 pKa = 8.43PRR13 pKa = 11.84KK14 pKa = 8.99KK15 pKa = 10.03VCGFCVDD22 pKa = 3.21KK23 pKa = 11.52VEE25 pKa = 4.75NIDD28 pKa = 3.57YY29 pKa = 10.94KK30 pKa = 11.15DD31 pKa = 3.08IARR34 pKa = 11.84LRR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 8.42MSEE41 pKa = 3.7RR42 pKa = 11.84GKK44 pKa = 10.14ILPRR48 pKa = 11.84RR49 pKa = 11.84VTGTCAAHH57 pKa = 5.98QRR59 pKa = 11.84ALTVAIKK66 pKa = 10.31RR67 pKa = 11.84ARR69 pKa = 11.84HH70 pKa = 5.37LALLPYY76 pKa = 8.42TTDD79 pKa = 2.85
Molecular weight: 9.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
812747 |
34 |
4072 |
324.1 |
36.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.074 ± 0.051 | 1.588 ± 0.025 |
5.872 ± 0.046 | 7.493 ± 0.05 |
4.359 ± 0.038 | 6.966 ± 0.046 |
1.471 ± 0.018 | 7.59 ± 0.044 |
7.288 ± 0.05 | 8.414 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.795 ± 0.025 | 5.087 ± 0.043 |
3.246 ± 0.028 | 2.856 ± 0.026 |
3.895 ± 0.041 | 6.426 ± 0.043 |
5.745 ± 0.05 | 6.858 ± 0.039 |
0.841 ± 0.018 | 4.137 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
