
Lake Sarah-associated circular virus-27
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.54
Get precalculated fractions of proteins
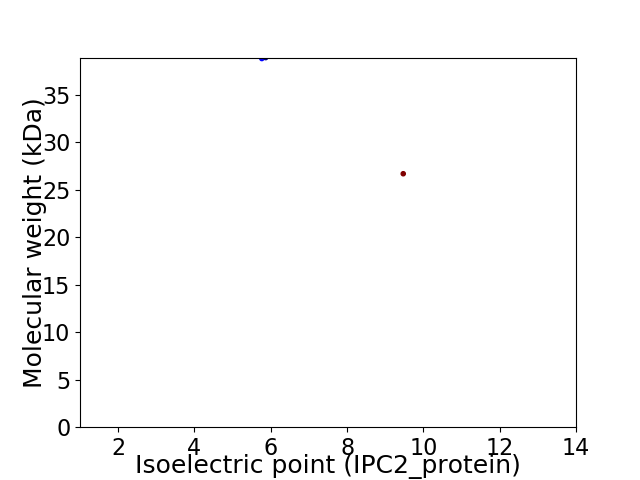
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQS3|A0A140AQS3_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-27 OX=1685754 PE=3 SV=1
MM1 pKa = 8.02DD2 pKa = 4.38PQGRR6 pKa = 11.84GWCFTLNNPDD16 pKa = 4.55DD17 pKa = 5.46LDD19 pKa = 3.82VLLPQSWDD27 pKa = 3.38PDD29 pKa = 3.82SYY31 pKa = 11.91DD32 pKa = 3.35YY33 pKa = 11.38LVYY36 pKa = 10.2HH37 pKa = 6.84LEE39 pKa = 4.02MGAEE43 pKa = 4.64GTAHH47 pKa = 6.03LQGYY51 pKa = 9.21LYY53 pKa = 10.24FGSRR57 pKa = 11.84IRR59 pKa = 11.84FSQLKK64 pKa = 9.2KK65 pKa = 8.57WFPGSRR71 pKa = 11.84VHH73 pKa = 7.86LEE75 pKa = 3.86AAKK78 pKa = 9.08GTAVQNQRR86 pKa = 11.84YY87 pKa = 6.1CTKK90 pKa = 10.69EE91 pKa = 3.67EE92 pKa = 4.14GRR94 pKa = 11.84LEE96 pKa = 4.31GPWEE100 pKa = 4.42FGSLPDD106 pKa = 3.7QSQGKK111 pKa = 10.08RR112 pKa = 11.84NDD114 pKa = 3.6LLAVKK119 pKa = 10.44ADD121 pKa = 3.99LDD123 pKa = 4.08AGASLLEE130 pKa = 4.18IADD133 pKa = 3.81DD134 pKa = 3.79HH135 pKa = 6.6FSAFVRR141 pKa = 11.84YY142 pKa = 8.79HH143 pKa = 7.0RR144 pKa = 11.84SFKK147 pKa = 10.4EE148 pKa = 3.83YY149 pKa = 10.55KK150 pKa = 9.86LLKK153 pKa = 10.72APVANVAKK161 pKa = 10.36KK162 pKa = 10.34IIVVWGPTGTGKK174 pKa = 8.17TRR176 pKa = 11.84WAFDD180 pKa = 3.64NYY182 pKa = 10.29PSAFWKK188 pKa = 9.1TKK190 pKa = 10.56SGGSQQFWDD199 pKa = 4.88GYY201 pKa = 10.49DD202 pKa = 3.6GEE204 pKa = 4.7SVIIVDD210 pKa = 5.04EE211 pKa = 4.88FYY213 pKa = 11.1GWLPFDD219 pKa = 3.9FMLRR223 pKa = 11.84FLDD226 pKa = 4.29RR227 pKa = 11.84YY228 pKa = 8.88PVKK231 pKa = 10.93LEE233 pKa = 4.05IKK235 pKa = 9.94HH236 pKa = 5.29GTVPLCADD244 pKa = 4.32TIIFTSNKK252 pKa = 9.39HH253 pKa = 4.83PSEE256 pKa = 4.11WYY258 pKa = 9.97HH259 pKa = 4.6IQRR262 pKa = 11.84YY263 pKa = 8.47KK264 pKa = 10.21WDD266 pKa = 3.41EE267 pKa = 4.21SNPLRR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84LTSVRR279 pKa = 11.84EE280 pKa = 3.79LRR282 pKa = 11.84ADD284 pKa = 3.31GFGPDD289 pKa = 3.42EE290 pKa = 4.52SSPGFSSSVAVVAPVFTTTTTTTTTSAGGLDD321 pKa = 3.57DD322 pKa = 4.02VPRR325 pKa = 11.84FRR327 pKa = 11.84NGYY330 pKa = 6.44PAPTVTDD337 pKa = 4.56LYY339 pKa = 11.47KK340 pKa = 10.94DD341 pKa = 3.47LL342 pKa = 5.36
MM1 pKa = 8.02DD2 pKa = 4.38PQGRR6 pKa = 11.84GWCFTLNNPDD16 pKa = 4.55DD17 pKa = 5.46LDD19 pKa = 3.82VLLPQSWDD27 pKa = 3.38PDD29 pKa = 3.82SYY31 pKa = 11.91DD32 pKa = 3.35YY33 pKa = 11.38LVYY36 pKa = 10.2HH37 pKa = 6.84LEE39 pKa = 4.02MGAEE43 pKa = 4.64GTAHH47 pKa = 6.03LQGYY51 pKa = 9.21LYY53 pKa = 10.24FGSRR57 pKa = 11.84IRR59 pKa = 11.84FSQLKK64 pKa = 9.2KK65 pKa = 8.57WFPGSRR71 pKa = 11.84VHH73 pKa = 7.86LEE75 pKa = 3.86AAKK78 pKa = 9.08GTAVQNQRR86 pKa = 11.84YY87 pKa = 6.1CTKK90 pKa = 10.69EE91 pKa = 3.67EE92 pKa = 4.14GRR94 pKa = 11.84LEE96 pKa = 4.31GPWEE100 pKa = 4.42FGSLPDD106 pKa = 3.7QSQGKK111 pKa = 10.08RR112 pKa = 11.84NDD114 pKa = 3.6LLAVKK119 pKa = 10.44ADD121 pKa = 3.99LDD123 pKa = 4.08AGASLLEE130 pKa = 4.18IADD133 pKa = 3.81DD134 pKa = 3.79HH135 pKa = 6.6FSAFVRR141 pKa = 11.84YY142 pKa = 8.79HH143 pKa = 7.0RR144 pKa = 11.84SFKK147 pKa = 10.4EE148 pKa = 3.83YY149 pKa = 10.55KK150 pKa = 9.86LLKK153 pKa = 10.72APVANVAKK161 pKa = 10.36KK162 pKa = 10.34IIVVWGPTGTGKK174 pKa = 8.17TRR176 pKa = 11.84WAFDD180 pKa = 3.64NYY182 pKa = 10.29PSAFWKK188 pKa = 9.1TKK190 pKa = 10.56SGGSQQFWDD199 pKa = 4.88GYY201 pKa = 10.49DD202 pKa = 3.6GEE204 pKa = 4.7SVIIVDD210 pKa = 5.04EE211 pKa = 4.88FYY213 pKa = 11.1GWLPFDD219 pKa = 3.9FMLRR223 pKa = 11.84FLDD226 pKa = 4.29RR227 pKa = 11.84YY228 pKa = 8.88PVKK231 pKa = 10.93LEE233 pKa = 4.05IKK235 pKa = 9.94HH236 pKa = 5.29GTVPLCADD244 pKa = 4.32TIIFTSNKK252 pKa = 9.39HH253 pKa = 4.83PSEE256 pKa = 4.11WYY258 pKa = 9.97HH259 pKa = 4.6IQRR262 pKa = 11.84YY263 pKa = 8.47KK264 pKa = 10.21WDD266 pKa = 3.41EE267 pKa = 4.21SNPLRR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84LTSVRR279 pKa = 11.84EE280 pKa = 3.79LRR282 pKa = 11.84ADD284 pKa = 3.31GFGPDD289 pKa = 3.42EE290 pKa = 4.52SSPGFSSSVAVVAPVFTTTTTTTTTSAGGLDD321 pKa = 3.57DD322 pKa = 4.02VPRR325 pKa = 11.84FRR327 pKa = 11.84NGYY330 pKa = 6.44PAPTVTDD337 pKa = 4.56LYY339 pKa = 11.47KK340 pKa = 10.94DD341 pKa = 3.47LL342 pKa = 5.36
Molecular weight: 38.81 kDa
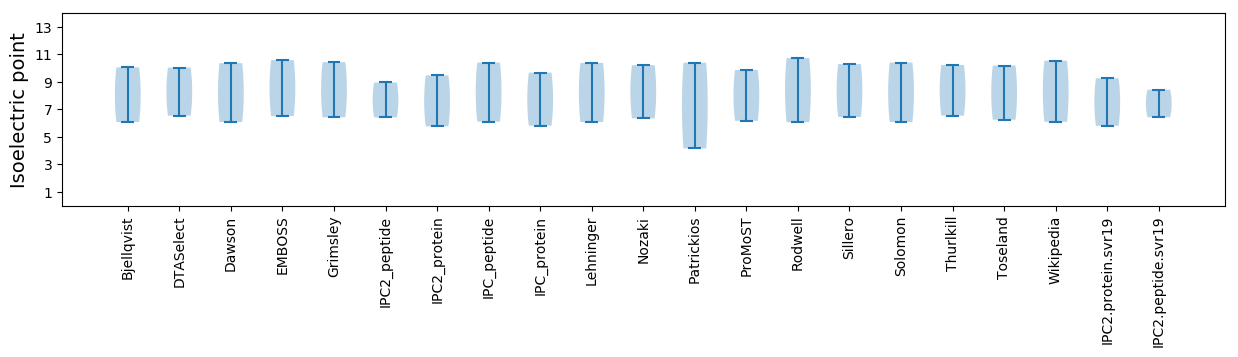
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQS3|A0A140AQS3_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-27 OX=1685754 PE=3 SV=1
MM1 pKa = 7.45PRR3 pKa = 11.84GSIFAKK9 pKa = 10.38KK10 pKa = 10.22SSTYY14 pKa = 9.25QKK16 pKa = 10.65RR17 pKa = 11.84KK18 pKa = 9.11LAAAAKK24 pKa = 9.57AISAARR30 pKa = 11.84GTTLSSRR37 pKa = 11.84RR38 pKa = 11.84GLSAPLRR45 pKa = 11.84TGGFWGQEE53 pKa = 3.17RR54 pKa = 11.84RR55 pKa = 11.84MYY57 pKa = 10.91SGMGPEE63 pKa = 4.78LKK65 pKa = 10.65VIDD68 pKa = 4.49SGPVGPLGFATAGAVALLNGVAQGTDD94 pKa = 3.04DD95 pKa = 3.48TQRR98 pKa = 11.84IGRR101 pKa = 11.84RR102 pKa = 11.84ATIKK106 pKa = 10.65SILFRR111 pKa = 11.84AFINPTTTTDD121 pKa = 4.03NIGDD125 pKa = 3.45QCRR128 pKa = 11.84FLLVWDD134 pKa = 3.93NQANGTALTVANVLNSATFSEE155 pKa = 4.84PNNLDD160 pKa = 2.89NRR162 pKa = 11.84DD163 pKa = 3.45RR164 pKa = 11.84FKK166 pKa = 10.88IIWDD170 pKa = 3.12KK171 pKa = 10.39WITMNPAVYY180 pKa = 9.75AANVFTGGNPQCKK193 pKa = 8.61MVHH196 pKa = 6.38CYY198 pKa = 10.37KK199 pKa = 10.66KK200 pKa = 10.97LNLEE204 pKa = 4.14TTFNGSGATIGSIATGSLYY223 pKa = 10.93VIAIAQNATNNNYY236 pKa = 9.75VYY238 pKa = 10.3NCRR241 pKa = 11.84VRR243 pKa = 11.84FTDD246 pKa = 3.09AA247 pKa = 3.91
MM1 pKa = 7.45PRR3 pKa = 11.84GSIFAKK9 pKa = 10.38KK10 pKa = 10.22SSTYY14 pKa = 9.25QKK16 pKa = 10.65RR17 pKa = 11.84KK18 pKa = 9.11LAAAAKK24 pKa = 9.57AISAARR30 pKa = 11.84GTTLSSRR37 pKa = 11.84RR38 pKa = 11.84GLSAPLRR45 pKa = 11.84TGGFWGQEE53 pKa = 3.17RR54 pKa = 11.84RR55 pKa = 11.84MYY57 pKa = 10.91SGMGPEE63 pKa = 4.78LKK65 pKa = 10.65VIDD68 pKa = 4.49SGPVGPLGFATAGAVALLNGVAQGTDD94 pKa = 3.04DD95 pKa = 3.48TQRR98 pKa = 11.84IGRR101 pKa = 11.84RR102 pKa = 11.84ATIKK106 pKa = 10.65SILFRR111 pKa = 11.84AFINPTTTTDD121 pKa = 4.03NIGDD125 pKa = 3.45QCRR128 pKa = 11.84FLLVWDD134 pKa = 3.93NQANGTALTVANVLNSATFSEE155 pKa = 4.84PNNLDD160 pKa = 2.89NRR162 pKa = 11.84DD163 pKa = 3.45RR164 pKa = 11.84FKK166 pKa = 10.88IIWDD170 pKa = 3.12KK171 pKa = 10.39WITMNPAVYY180 pKa = 9.75AANVFTGGNPQCKK193 pKa = 8.61MVHH196 pKa = 6.38CYY198 pKa = 10.37KK199 pKa = 10.66KK200 pKa = 10.97LNLEE204 pKa = 4.14TTFNGSGATIGSIATGSLYY223 pKa = 10.93VIAIAQNATNNNYY236 pKa = 9.75VYY238 pKa = 10.3NCRR241 pKa = 11.84VRR243 pKa = 11.84FTDD246 pKa = 3.09AA247 pKa = 3.91
Molecular weight: 26.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
589 |
247 |
342 |
294.5 |
32.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.659 ± 1.717 | 1.188 ± 0.24 |
6.282 ± 1.244 | 3.396 ± 0.989 |
5.263 ± 0.451 | 8.659 ± 0.364 |
1.528 ± 0.626 | 4.244 ± 1.019 |
5.263 ± 0.226 | 7.98 ± 0.611 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.358 ± 0.371 | 4.924 ± 1.768 |
5.093 ± 0.808 | 3.226 ± 0.007 |
6.282 ± 0.335 | 6.621 ± 0.306 |
7.81 ± 0.837 | 5.772 ± 0.284 |
2.547 ± 0.517 | 3.905 ± 0.597 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
