
Oceanispirochaeta sp. M1
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Oceanispirochaeta; unclassified Oceanispirochaeta
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
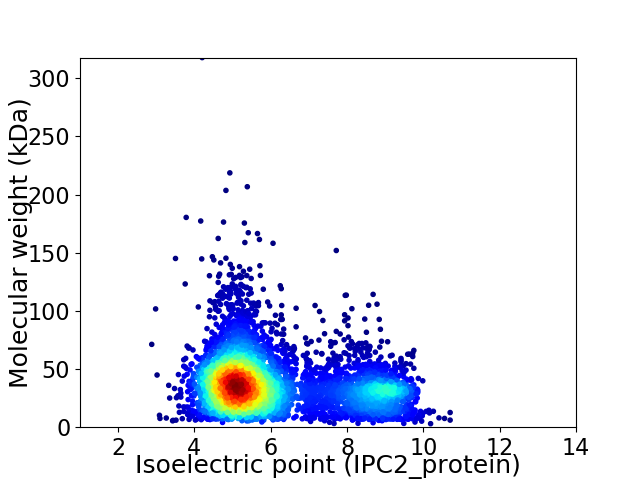
Virtual 2D-PAGE plot for 5141 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370ARX4|A0A370ARX4_9SPIO LacI family transcriptional regulator OS=Oceanispirochaeta sp. M1 OX=2283433 GN=DV872_09130 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 9.34KK3 pKa = 9.12TIVLLTIVLALVAAPVFAEE22 pKa = 4.17TGFTGSVEE30 pKa = 4.37FKK32 pKa = 10.73YY33 pKa = 9.83GTDD36 pKa = 3.66FSDD39 pKa = 4.18FATDD43 pKa = 3.2TGNDD47 pKa = 3.49TKK49 pKa = 10.85ISLDD53 pKa = 3.47GKK55 pKa = 9.89VGEE58 pKa = 4.76FSTISVGIEE67 pKa = 3.36ADD69 pKa = 3.69EE70 pKa = 4.43NSNASATLMTLSQDD84 pKa = 3.22VTGALGVEE92 pKa = 4.8GPVSFAYY99 pKa = 9.78KK100 pKa = 9.48MGKK103 pKa = 7.41QTYY106 pKa = 9.2QPADD110 pKa = 3.36YY111 pKa = 10.21SGVGGYY117 pKa = 10.31EE118 pKa = 3.83DD119 pKa = 4.15AGRR122 pKa = 11.84DD123 pKa = 3.48AKK125 pKa = 10.56IGQGYY130 pKa = 8.36EE131 pKa = 3.92ADD133 pKa = 3.86DD134 pKa = 3.83TDD136 pKa = 3.82FTTNSMLGTVLTFGFADD153 pKa = 4.42MIMIDD158 pKa = 3.53AVVFPASYY166 pKa = 10.39FEE168 pKa = 4.36KK169 pKa = 10.83AADD172 pKa = 3.51AANGYY177 pKa = 10.17FDD179 pKa = 5.42ADD181 pKa = 3.51NFAEE185 pKa = 4.48FAVNAYY191 pKa = 7.88GTFGPAQVSVYY202 pKa = 8.65YY203 pKa = 10.61TMSNLSYY210 pKa = 10.72LADD213 pKa = 3.8SDD215 pKa = 4.22GDD217 pKa = 4.52AIDD220 pKa = 4.2EE221 pKa = 4.54AGDD224 pKa = 3.73MIGANLGLVIDD235 pKa = 4.39ALSVGVLWEE244 pKa = 4.04TDD246 pKa = 3.45LEE248 pKa = 4.43KK249 pKa = 11.04DD250 pKa = 3.55FQAATGVTGTKK261 pKa = 10.05PMNTEE266 pKa = 3.04IGIAAKK272 pKa = 10.26YY273 pKa = 10.13VIGSVTPGIAFKK285 pKa = 9.79TAFYY289 pKa = 10.65DD290 pKa = 4.15GSDD293 pKa = 3.67FAADD297 pKa = 3.36SGLGLNVTWAAAEE310 pKa = 4.14MFNVTAAVKK319 pKa = 10.55NSFVFDD325 pKa = 4.32DD326 pKa = 4.69LGDD329 pKa = 3.69TLGYY333 pKa = 10.07EE334 pKa = 4.11VGVDD338 pKa = 3.49YY339 pKa = 10.8TLDD342 pKa = 3.39GVKK345 pKa = 10.63YY346 pKa = 10.59SIGYY350 pKa = 8.01TDD352 pKa = 3.53GSEE355 pKa = 4.34FKK357 pKa = 10.69AYY359 pKa = 10.07DD360 pKa = 3.78AEE362 pKa = 5.12LSDD365 pKa = 3.84TGNMYY370 pKa = 10.84VKK372 pKa = 10.57VGASFF377 pKa = 3.67
MM1 pKa = 7.66KK2 pKa = 9.34KK3 pKa = 9.12TIVLLTIVLALVAAPVFAEE22 pKa = 4.17TGFTGSVEE30 pKa = 4.37FKK32 pKa = 10.73YY33 pKa = 9.83GTDD36 pKa = 3.66FSDD39 pKa = 4.18FATDD43 pKa = 3.2TGNDD47 pKa = 3.49TKK49 pKa = 10.85ISLDD53 pKa = 3.47GKK55 pKa = 9.89VGEE58 pKa = 4.76FSTISVGIEE67 pKa = 3.36ADD69 pKa = 3.69EE70 pKa = 4.43NSNASATLMTLSQDD84 pKa = 3.22VTGALGVEE92 pKa = 4.8GPVSFAYY99 pKa = 9.78KK100 pKa = 9.48MGKK103 pKa = 7.41QTYY106 pKa = 9.2QPADD110 pKa = 3.36YY111 pKa = 10.21SGVGGYY117 pKa = 10.31EE118 pKa = 3.83DD119 pKa = 4.15AGRR122 pKa = 11.84DD123 pKa = 3.48AKK125 pKa = 10.56IGQGYY130 pKa = 8.36EE131 pKa = 3.92ADD133 pKa = 3.86DD134 pKa = 3.83TDD136 pKa = 3.82FTTNSMLGTVLTFGFADD153 pKa = 4.42MIMIDD158 pKa = 3.53AVVFPASYY166 pKa = 10.39FEE168 pKa = 4.36KK169 pKa = 10.83AADD172 pKa = 3.51AANGYY177 pKa = 10.17FDD179 pKa = 5.42ADD181 pKa = 3.51NFAEE185 pKa = 4.48FAVNAYY191 pKa = 7.88GTFGPAQVSVYY202 pKa = 8.65YY203 pKa = 10.61TMSNLSYY210 pKa = 10.72LADD213 pKa = 3.8SDD215 pKa = 4.22GDD217 pKa = 4.52AIDD220 pKa = 4.2EE221 pKa = 4.54AGDD224 pKa = 3.73MIGANLGLVIDD235 pKa = 4.39ALSVGVLWEE244 pKa = 4.04TDD246 pKa = 3.45LEE248 pKa = 4.43KK249 pKa = 11.04DD250 pKa = 3.55FQAATGVTGTKK261 pKa = 10.05PMNTEE266 pKa = 3.04IGIAAKK272 pKa = 10.26YY273 pKa = 10.13VIGSVTPGIAFKK285 pKa = 9.79TAFYY289 pKa = 10.65DD290 pKa = 4.15GSDD293 pKa = 3.67FAADD297 pKa = 3.36SGLGLNVTWAAAEE310 pKa = 4.14MFNVTAAVKK319 pKa = 10.55NSFVFDD325 pKa = 4.32DD326 pKa = 4.69LGDD329 pKa = 3.69TLGYY333 pKa = 10.07EE334 pKa = 4.11VGVDD338 pKa = 3.49YY339 pKa = 10.8TLDD342 pKa = 3.39GVKK345 pKa = 10.63YY346 pKa = 10.59SIGYY350 pKa = 8.01TDD352 pKa = 3.53GSEE355 pKa = 4.34FKK357 pKa = 10.69AYY359 pKa = 10.07DD360 pKa = 3.78AEE362 pKa = 5.12LSDD365 pKa = 3.84TGNMYY370 pKa = 10.84VKK372 pKa = 10.57VGASFF377 pKa = 3.67
Molecular weight: 39.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370AUF5|A0A370AUF5_9SPIO HAMP domain-containing protein OS=Oceanispirochaeta sp. M1 OX=2283433 GN=DV872_12745 PE=4 SV=1
MM1 pKa = 7.62AKK3 pKa = 10.23KK4 pKa = 10.87SMIVKK9 pKa = 10.12CNRR12 pKa = 11.84KK13 pKa = 8.44QKK15 pKa = 10.53YY16 pKa = 6.66STRR19 pKa = 11.84EE20 pKa = 3.65YY21 pKa = 10.59NRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PRR31 pKa = 11.84GYY33 pKa = 8.93MRR35 pKa = 11.84KK36 pKa = 9.48FEE38 pKa = 3.99MCRR41 pKa = 11.84ICFRR45 pKa = 11.84KK46 pKa = 9.57LANEE50 pKa = 3.91GLIPGVTKK58 pKa = 10.8SSWW61 pKa = 2.87
MM1 pKa = 7.62AKK3 pKa = 10.23KK4 pKa = 10.87SMIVKK9 pKa = 10.12CNRR12 pKa = 11.84KK13 pKa = 8.44QKK15 pKa = 10.53YY16 pKa = 6.66STRR19 pKa = 11.84EE20 pKa = 3.65YY21 pKa = 10.59NRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PRR31 pKa = 11.84GYY33 pKa = 8.93MRR35 pKa = 11.84KK36 pKa = 9.48FEE38 pKa = 3.99MCRR41 pKa = 11.84ICFRR45 pKa = 11.84KK46 pKa = 9.57LANEE50 pKa = 3.91GLIPGVTKK58 pKa = 10.8SSWW61 pKa = 2.87
Molecular weight: 7.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1776840 |
27 |
2815 |
345.6 |
38.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.584 ± 0.038 | 1.011 ± 0.011 |
5.812 ± 0.027 | 7.163 ± 0.039 |
4.687 ± 0.023 | 6.978 ± 0.033 |
1.782 ± 0.014 | 7.804 ± 0.036 |
6.122 ± 0.03 | 10.358 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.883 ± 0.018 | 4.394 ± 0.025 |
3.829 ± 0.019 | 3.039 ± 0.017 |
4.636 ± 0.025 | 7.43 ± 0.028 |
4.861 ± 0.021 | 5.915 ± 0.027 |
1.134 ± 0.012 | 3.577 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
