
Fusarium poae virus 1-240374
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus; Fusarium poae virus 1
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
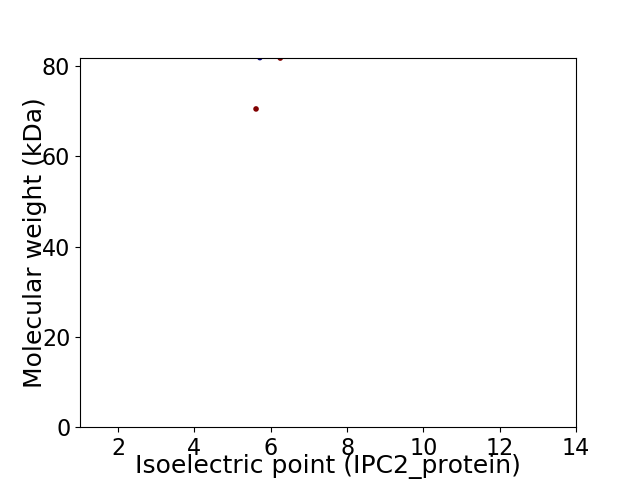
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B4ZA50|A0A1B4ZA50_9VIRU RNA-dependent RNA polymerase OS=Fusarium poae virus 1-240374 OX=1849534 PE=4 SV=1
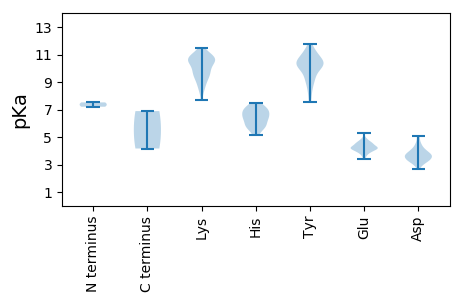
MM1 pKa = 7.2SQQNNARR8 pKa = 11.84VSTASRR14 pKa = 11.84TSSVEE19 pKa = 3.41LRR21 pKa = 11.84TPAAIAAIAKK31 pKa = 9.59RR32 pKa = 11.84FAPVVFQTDD41 pKa = 3.83DD42 pKa = 3.77TPVPPSNADD51 pKa = 2.94YY52 pKa = 9.79FARR55 pKa = 11.84MNMPSNIEE63 pKa = 4.18LDD65 pKa = 3.6SGDD68 pKa = 3.91YY69 pKa = 9.97ATIDD73 pKa = 3.42VQFDD77 pKa = 3.17TRR79 pKa = 11.84FIMNYY84 pKa = 9.55ILYY87 pKa = 9.28HH88 pKa = 5.16ATNRR92 pKa = 11.84APEE95 pKa = 4.2MNFKK99 pKa = 9.78GHH101 pKa = 7.34PYY103 pKa = 8.65ATPLSYY109 pKa = 10.79AGYY112 pKa = 10.11CLTLLYY118 pKa = 10.69TFMLAVDD125 pKa = 3.69VTTRR129 pKa = 11.84PEE131 pKa = 4.3KK132 pKa = 9.75SWHH135 pKa = 5.87AARR138 pKa = 11.84FMSDD142 pKa = 3.07AEE144 pKa = 4.27RR145 pKa = 11.84KK146 pKa = 9.65DD147 pKa = 3.75LYY149 pKa = 11.15DD150 pKa = 3.22VLLSSRR156 pKa = 11.84VPTWFVDD163 pKa = 3.67LLIEE167 pKa = 4.18FAPVFDD173 pKa = 4.12PRR175 pKa = 11.84RR176 pKa = 11.84NNILMVPTLAGYY188 pKa = 9.0IHH190 pKa = 6.96NIDD193 pKa = 4.0FGRR196 pKa = 11.84TFPPSIFYY204 pKa = 10.4SAHH207 pKa = 6.03NLLASTRR214 pKa = 11.84TNLDD218 pKa = 3.26PDD220 pKa = 3.83NVIDD224 pKa = 4.54SLMGTPLINHH234 pKa = 6.41GTNAYY239 pKa = 7.58TVSNYY244 pKa = 10.67LGTWYY249 pKa = 10.11DD250 pKa = 3.63AGHH253 pKa = 6.8HH254 pKa = 6.4PNFVNQDD261 pKa = 3.28YY262 pKa = 9.77MSFFNPLVGRR272 pKa = 11.84FLTQRR277 pKa = 11.84PTFARR282 pKa = 11.84LPFSPEE288 pKa = 3.59DD289 pKa = 3.54LADD292 pKa = 3.71NGIGNIYY299 pKa = 7.68TTFLLASDD307 pKa = 4.47EE308 pKa = 4.65NISLANTLYY317 pKa = 10.13TAMSTFILSDD327 pKa = 3.67EE328 pKa = 4.58PNAPQLGSVLSSLSGSLLLSHH349 pKa = 7.09SIEE352 pKa = 5.29PITLPTWTKK361 pKa = 8.58ATYY364 pKa = 10.64VSDD367 pKa = 3.59IDD369 pKa = 3.88PSLVSDD375 pKa = 3.73SKK377 pKa = 11.14FVSDD381 pKa = 4.17HH382 pKa = 6.2NLFSRR387 pKa = 11.84TPKK390 pKa = 10.25YY391 pKa = 10.38EE392 pKa = 3.88KK393 pKa = 9.85STLFPNDD400 pKa = 2.99GKK402 pKa = 9.5TITSLYY408 pKa = 10.5YY409 pKa = 10.53LVTKK413 pKa = 10.27KK414 pKa = 10.56KK415 pKa = 10.02HH416 pKa = 6.15DD417 pKa = 4.31ANDD420 pKa = 3.43NPTSHH425 pKa = 7.23LSFKK429 pKa = 10.9GKK431 pKa = 10.38DD432 pKa = 3.24SVTPYY437 pKa = 10.98VLYY440 pKa = 9.31FQPYY444 pKa = 8.43DD445 pKa = 3.62VSPSSLGLTIAAGLKK460 pKa = 9.86IEE462 pKa = 4.23LAEE465 pKa = 3.93ISGFTVPVEE474 pKa = 4.18HH475 pKa = 7.41PEE477 pKa = 4.87SSLLDD482 pKa = 3.63NNSQYY487 pKa = 10.91LQSAIRR493 pKa = 11.84ANLIAPVNDD502 pKa = 3.24RR503 pKa = 11.84STAAEE508 pKa = 3.84NSSLNLARR516 pKa = 11.84SGLDD520 pKa = 2.98STLQGITASFISTCKK535 pKa = 10.26SVLPFFDD542 pKa = 4.57NEE544 pKa = 4.22GNNLGDD550 pKa = 3.73STDD553 pKa = 3.37LSKK556 pKa = 11.2FGLTPEE562 pKa = 4.28LHH564 pKa = 6.51HH565 pKa = 6.43VSPFAGFNVKK575 pKa = 10.33AGTNGQLLSTEE586 pKa = 4.18KK587 pKa = 10.64SIHH590 pKa = 5.69LWSSYY595 pKa = 10.17RR596 pKa = 11.84VVHH599 pKa = 5.64TKK601 pKa = 10.43KK602 pKa = 10.58NPAARR607 pKa = 11.84DD608 pKa = 3.29ISFIASFRR616 pKa = 11.84PIYY619 pKa = 8.34GTNVTLSRR627 pKa = 11.84SKK629 pKa = 10.85NPSLIIPHH637 pKa = 6.89
MM1 pKa = 7.2SQQNNARR8 pKa = 11.84VSTASRR14 pKa = 11.84TSSVEE19 pKa = 3.41LRR21 pKa = 11.84TPAAIAAIAKK31 pKa = 9.59RR32 pKa = 11.84FAPVVFQTDD41 pKa = 3.83DD42 pKa = 3.77TPVPPSNADD51 pKa = 2.94YY52 pKa = 9.79FARR55 pKa = 11.84MNMPSNIEE63 pKa = 4.18LDD65 pKa = 3.6SGDD68 pKa = 3.91YY69 pKa = 9.97ATIDD73 pKa = 3.42VQFDD77 pKa = 3.17TRR79 pKa = 11.84FIMNYY84 pKa = 9.55ILYY87 pKa = 9.28HH88 pKa = 5.16ATNRR92 pKa = 11.84APEE95 pKa = 4.2MNFKK99 pKa = 9.78GHH101 pKa = 7.34PYY103 pKa = 8.65ATPLSYY109 pKa = 10.79AGYY112 pKa = 10.11CLTLLYY118 pKa = 10.69TFMLAVDD125 pKa = 3.69VTTRR129 pKa = 11.84PEE131 pKa = 4.3KK132 pKa = 9.75SWHH135 pKa = 5.87AARR138 pKa = 11.84FMSDD142 pKa = 3.07AEE144 pKa = 4.27RR145 pKa = 11.84KK146 pKa = 9.65DD147 pKa = 3.75LYY149 pKa = 11.15DD150 pKa = 3.22VLLSSRR156 pKa = 11.84VPTWFVDD163 pKa = 3.67LLIEE167 pKa = 4.18FAPVFDD173 pKa = 4.12PRR175 pKa = 11.84RR176 pKa = 11.84NNILMVPTLAGYY188 pKa = 9.0IHH190 pKa = 6.96NIDD193 pKa = 4.0FGRR196 pKa = 11.84TFPPSIFYY204 pKa = 10.4SAHH207 pKa = 6.03NLLASTRR214 pKa = 11.84TNLDD218 pKa = 3.26PDD220 pKa = 3.83NVIDD224 pKa = 4.54SLMGTPLINHH234 pKa = 6.41GTNAYY239 pKa = 7.58TVSNYY244 pKa = 10.67LGTWYY249 pKa = 10.11DD250 pKa = 3.63AGHH253 pKa = 6.8HH254 pKa = 6.4PNFVNQDD261 pKa = 3.28YY262 pKa = 9.77MSFFNPLVGRR272 pKa = 11.84FLTQRR277 pKa = 11.84PTFARR282 pKa = 11.84LPFSPEE288 pKa = 3.59DD289 pKa = 3.54LADD292 pKa = 3.71NGIGNIYY299 pKa = 7.68TTFLLASDD307 pKa = 4.47EE308 pKa = 4.65NISLANTLYY317 pKa = 10.13TAMSTFILSDD327 pKa = 3.67EE328 pKa = 4.58PNAPQLGSVLSSLSGSLLLSHH349 pKa = 7.09SIEE352 pKa = 5.29PITLPTWTKK361 pKa = 8.58ATYY364 pKa = 10.64VSDD367 pKa = 3.59IDD369 pKa = 3.88PSLVSDD375 pKa = 3.73SKK377 pKa = 11.14FVSDD381 pKa = 4.17HH382 pKa = 6.2NLFSRR387 pKa = 11.84TPKK390 pKa = 10.25YY391 pKa = 10.38EE392 pKa = 3.88KK393 pKa = 9.85STLFPNDD400 pKa = 2.99GKK402 pKa = 9.5TITSLYY408 pKa = 10.5YY409 pKa = 10.53LVTKK413 pKa = 10.27KK414 pKa = 10.56KK415 pKa = 10.02HH416 pKa = 6.15DD417 pKa = 4.31ANDD420 pKa = 3.43NPTSHH425 pKa = 7.23LSFKK429 pKa = 10.9GKK431 pKa = 10.38DD432 pKa = 3.24SVTPYY437 pKa = 10.98VLYY440 pKa = 9.31FQPYY444 pKa = 8.43DD445 pKa = 3.62VSPSSLGLTIAAGLKK460 pKa = 9.86IEE462 pKa = 4.23LAEE465 pKa = 3.93ISGFTVPVEE474 pKa = 4.18HH475 pKa = 7.41PEE477 pKa = 4.87SSLLDD482 pKa = 3.63NNSQYY487 pKa = 10.91LQSAIRR493 pKa = 11.84ANLIAPVNDD502 pKa = 3.24RR503 pKa = 11.84STAAEE508 pKa = 3.84NSSLNLARR516 pKa = 11.84SGLDD520 pKa = 2.98STLQGITASFISTCKK535 pKa = 10.26SVLPFFDD542 pKa = 4.57NEE544 pKa = 4.22GNNLGDD550 pKa = 3.73STDD553 pKa = 3.37LSKK556 pKa = 11.2FGLTPEE562 pKa = 4.28LHH564 pKa = 6.51HH565 pKa = 6.43VSPFAGFNVKK575 pKa = 10.33AGTNGQLLSTEE586 pKa = 4.18KK587 pKa = 10.64SIHH590 pKa = 5.69LWSSYY595 pKa = 10.17RR596 pKa = 11.84VVHH599 pKa = 5.64TKK601 pKa = 10.43KK602 pKa = 10.58NPAARR607 pKa = 11.84DD608 pKa = 3.29ISFIASFRR616 pKa = 11.84PIYY619 pKa = 8.34GTNVTLSRR627 pKa = 11.84SKK629 pKa = 10.85NPSLIIPHH637 pKa = 6.89
Molecular weight: 70.46 kDa
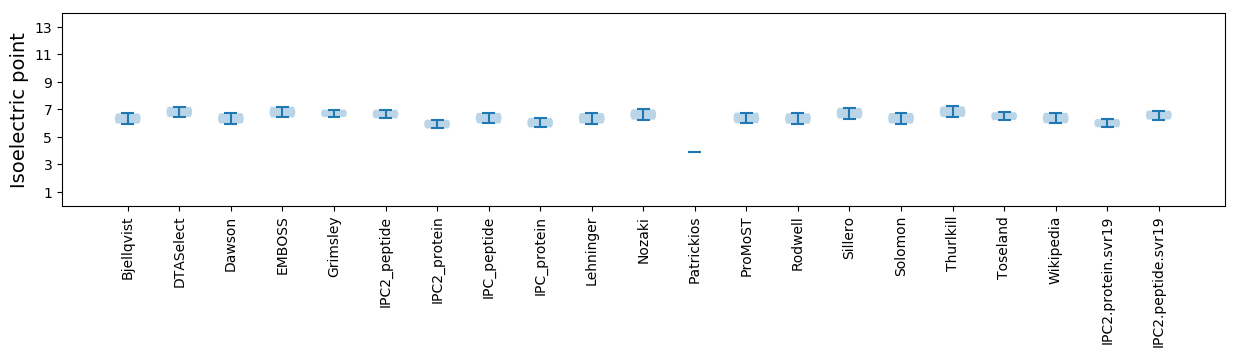
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B4ZA50|A0A1B4ZA50_9VIRU RNA-dependent RNA polymerase OS=Fusarium poae virus 1-240374 OX=1849534 PE=4 SV=1
MM1 pKa = 7.52LANIRR6 pKa = 11.84DD7 pKa = 3.95YY8 pKa = 11.13FHH10 pKa = 7.4EE11 pKa = 4.44KK12 pKa = 9.0LTRR15 pKa = 11.84LLYY18 pKa = 9.99DD19 pKa = 3.26HH20 pKa = 7.49KK21 pKa = 10.78IFQSNSKK28 pKa = 11.0DD29 pKa = 3.28PDD31 pKa = 3.39LTLEE35 pKa = 4.13AHH37 pKa = 6.56HH38 pKa = 6.79SSDD41 pKa = 3.21IEE43 pKa = 4.12RR44 pKa = 11.84IYY46 pKa = 11.22KK47 pKa = 9.85SIHH50 pKa = 5.92YY51 pKa = 9.6DD52 pKa = 3.59FNRR55 pKa = 11.84SPAPTDD61 pKa = 3.39YY62 pKa = 10.42EE63 pKa = 3.96AQYY66 pKa = 11.49QSIKK70 pKa = 10.51HH71 pKa = 5.63ILEE74 pKa = 4.86DD75 pKa = 3.63KK76 pKa = 10.58QSQQGFPHH84 pKa = 7.08EE85 pKa = 4.43YY86 pKa = 9.97YY87 pKa = 10.25RR88 pKa = 11.84LHH90 pKa = 7.04EE91 pKa = 4.51SPIPDD96 pKa = 3.99DD97 pKa = 5.1RR98 pKa = 11.84IPPSGIKK105 pKa = 10.19LLPFEE110 pKa = 4.54YY111 pKa = 10.56KK112 pKa = 10.65SMNVVTATPEE122 pKa = 4.01VPEE125 pKa = 4.58SGFKK129 pKa = 9.18IHH131 pKa = 6.9PRR133 pKa = 11.84IEE135 pKa = 3.62RR136 pKa = 11.84LLRR139 pKa = 11.84SKK141 pKa = 10.78YY142 pKa = 9.3PQYY145 pKa = 10.38LQYY148 pKa = 10.14VRR150 pKa = 11.84KK151 pKa = 7.66YY152 pKa = 8.1TRR154 pKa = 11.84PLGTTNATVSDD165 pKa = 4.28FFKK168 pKa = 10.55PQTPSQPVEE177 pKa = 3.79PTRR180 pKa = 11.84INHH183 pKa = 5.16VMSHH187 pKa = 5.22VMKK190 pKa = 10.8KK191 pKa = 8.65MAITPYY197 pKa = 10.55LPLHH201 pKa = 6.2FVDD204 pKa = 3.53TQYY207 pKa = 11.79DD208 pKa = 3.77KK209 pKa = 11.49RR210 pKa = 11.84PLANGTGYY218 pKa = 10.71HH219 pKa = 5.67NRR221 pKa = 11.84RR222 pKa = 11.84SHH224 pKa = 5.82EE225 pKa = 4.14MNIHH229 pKa = 6.49ALFSHH234 pKa = 6.39PKK236 pKa = 8.32EE237 pKa = 4.24YY238 pKa = 10.4EE239 pKa = 3.87SKK241 pKa = 8.84RR242 pKa = 11.84TSKK245 pKa = 10.71GYY247 pKa = 9.76YY248 pKa = 9.12VNAFLEE254 pKa = 4.58SARR257 pKa = 11.84SLIHH261 pKa = 6.68WIKK264 pKa = 10.88LYY266 pKa = 10.97GNPFRR271 pKa = 11.84HH272 pKa = 6.43CPSDD276 pKa = 3.31LAQSLRR282 pKa = 11.84EE283 pKa = 4.03FFLQRR288 pKa = 11.84PTMLFTRR295 pKa = 11.84NHH297 pKa = 5.57ISDD300 pKa = 3.52RR301 pKa = 11.84DD302 pKa = 4.03GILKK306 pKa = 9.32QRR308 pKa = 11.84PVYY311 pKa = 10.63AVDD314 pKa = 5.09DD315 pKa = 4.18LFLTIEE321 pKa = 4.17SMLTFPAHH329 pKa = 5.69VIARR333 pKa = 11.84KK334 pKa = 9.29PEE336 pKa = 3.9CCIMYY341 pKa = 10.3GLEE344 pKa = 4.27TIRR347 pKa = 11.84GSNQILDD354 pKa = 4.81KK355 pKa = 10.76IASDD359 pKa = 3.62YY360 pKa = 11.53KK361 pKa = 11.26SFFTIDD367 pKa = 2.65WSGFDD372 pKa = 4.51QRR374 pKa = 11.84LPWVIVKK381 pKa = 9.66LFFTEE386 pKa = 4.23YY387 pKa = 9.97IPRR390 pKa = 11.84LLVVNHH396 pKa = 6.92GYY398 pKa = 11.03APTYY402 pKa = 9.02EE403 pKa = 4.25YY404 pKa = 10.34PSYY407 pKa = 10.98PDD409 pKa = 3.35LTTNDD414 pKa = 3.23MVSRR418 pKa = 11.84LTNLLTFLATWYY430 pKa = 10.46FNMVFVTADD439 pKa = 2.87GFSYY443 pKa = 10.54VRR445 pKa = 11.84EE446 pKa = 4.19HH447 pKa = 6.82AGVPSGMLNTQFLDD461 pKa = 3.44SFGNLFLLIDD471 pKa = 3.82GLIEE475 pKa = 4.54FGSTDD480 pKa = 4.17AEE482 pKa = 4.06IDD484 pKa = 4.38DD485 pKa = 3.94ILLFIMGDD493 pKa = 3.62DD494 pKa = 3.56NSAFTTWSITHH505 pKa = 7.0LEE507 pKa = 4.04QFVSFFEE514 pKa = 4.66TYY516 pKa = 10.23ALSRR520 pKa = 11.84YY521 pKa = 9.99GMVLSKK527 pKa = 9.46TKK529 pKa = 10.69SIITTLRR536 pKa = 11.84HH537 pKa = 5.96KK538 pKa = 10.54IEE540 pKa = 4.17TLSYY544 pKa = 9.4QCNFGHH550 pKa = 7.22PRR552 pKa = 11.84RR553 pKa = 11.84PIGKK557 pKa = 9.07LVAQLCFPEE566 pKa = 4.83RR567 pKa = 11.84GPRR570 pKa = 11.84PKK572 pKa = 10.75YY573 pKa = 8.73MSARR577 pKa = 11.84AVGMAWASCGQDD589 pKa = 2.84KK590 pKa = 9.23TFHH593 pKa = 6.55DD594 pKa = 4.92FCRR597 pKa = 11.84DD598 pKa = 3.13VYY600 pKa = 11.3HH601 pKa = 6.92EE602 pKa = 4.75FNDD605 pKa = 4.1DD606 pKa = 3.93RR607 pKa = 11.84ADD609 pKa = 4.03LDD611 pKa = 3.62EE612 pKa = 4.4SAYY615 pKa = 11.34LHH617 pKa = 6.24IQSHH621 pKa = 6.05LPGYY625 pKa = 10.41LKK627 pKa = 9.66IDD629 pKa = 3.35EE630 pKa = 4.52SVRR633 pKa = 11.84QIVDD637 pKa = 3.74FQVFPSQQTVYY648 pKa = 9.09HH649 pKa = 5.79TVSRR653 pKa = 11.84WKK655 pKa = 10.9GPLSYY660 pKa = 10.33QPKK663 pKa = 8.42WDD665 pKa = 3.55LAHH668 pKa = 6.9FVNQPDD674 pKa = 3.99VVPPDD679 pKa = 3.4SVTLYY684 pKa = 10.05EE685 pKa = 4.5VWSEE689 pKa = 4.44LNFTPPILHH698 pKa = 6.6NLFF701 pKa = 4.17
MM1 pKa = 7.52LANIRR6 pKa = 11.84DD7 pKa = 3.95YY8 pKa = 11.13FHH10 pKa = 7.4EE11 pKa = 4.44KK12 pKa = 9.0LTRR15 pKa = 11.84LLYY18 pKa = 9.99DD19 pKa = 3.26HH20 pKa = 7.49KK21 pKa = 10.78IFQSNSKK28 pKa = 11.0DD29 pKa = 3.28PDD31 pKa = 3.39LTLEE35 pKa = 4.13AHH37 pKa = 6.56HH38 pKa = 6.79SSDD41 pKa = 3.21IEE43 pKa = 4.12RR44 pKa = 11.84IYY46 pKa = 11.22KK47 pKa = 9.85SIHH50 pKa = 5.92YY51 pKa = 9.6DD52 pKa = 3.59FNRR55 pKa = 11.84SPAPTDD61 pKa = 3.39YY62 pKa = 10.42EE63 pKa = 3.96AQYY66 pKa = 11.49QSIKK70 pKa = 10.51HH71 pKa = 5.63ILEE74 pKa = 4.86DD75 pKa = 3.63KK76 pKa = 10.58QSQQGFPHH84 pKa = 7.08EE85 pKa = 4.43YY86 pKa = 9.97YY87 pKa = 10.25RR88 pKa = 11.84LHH90 pKa = 7.04EE91 pKa = 4.51SPIPDD96 pKa = 3.99DD97 pKa = 5.1RR98 pKa = 11.84IPPSGIKK105 pKa = 10.19LLPFEE110 pKa = 4.54YY111 pKa = 10.56KK112 pKa = 10.65SMNVVTATPEE122 pKa = 4.01VPEE125 pKa = 4.58SGFKK129 pKa = 9.18IHH131 pKa = 6.9PRR133 pKa = 11.84IEE135 pKa = 3.62RR136 pKa = 11.84LLRR139 pKa = 11.84SKK141 pKa = 10.78YY142 pKa = 9.3PQYY145 pKa = 10.38LQYY148 pKa = 10.14VRR150 pKa = 11.84KK151 pKa = 7.66YY152 pKa = 8.1TRR154 pKa = 11.84PLGTTNATVSDD165 pKa = 4.28FFKK168 pKa = 10.55PQTPSQPVEE177 pKa = 3.79PTRR180 pKa = 11.84INHH183 pKa = 5.16VMSHH187 pKa = 5.22VMKK190 pKa = 10.8KK191 pKa = 8.65MAITPYY197 pKa = 10.55LPLHH201 pKa = 6.2FVDD204 pKa = 3.53TQYY207 pKa = 11.79DD208 pKa = 3.77KK209 pKa = 11.49RR210 pKa = 11.84PLANGTGYY218 pKa = 10.71HH219 pKa = 5.67NRR221 pKa = 11.84RR222 pKa = 11.84SHH224 pKa = 5.82EE225 pKa = 4.14MNIHH229 pKa = 6.49ALFSHH234 pKa = 6.39PKK236 pKa = 8.32EE237 pKa = 4.24YY238 pKa = 10.4EE239 pKa = 3.87SKK241 pKa = 8.84RR242 pKa = 11.84TSKK245 pKa = 10.71GYY247 pKa = 9.76YY248 pKa = 9.12VNAFLEE254 pKa = 4.58SARR257 pKa = 11.84SLIHH261 pKa = 6.68WIKK264 pKa = 10.88LYY266 pKa = 10.97GNPFRR271 pKa = 11.84HH272 pKa = 6.43CPSDD276 pKa = 3.31LAQSLRR282 pKa = 11.84EE283 pKa = 4.03FFLQRR288 pKa = 11.84PTMLFTRR295 pKa = 11.84NHH297 pKa = 5.57ISDD300 pKa = 3.52RR301 pKa = 11.84DD302 pKa = 4.03GILKK306 pKa = 9.32QRR308 pKa = 11.84PVYY311 pKa = 10.63AVDD314 pKa = 5.09DD315 pKa = 4.18LFLTIEE321 pKa = 4.17SMLTFPAHH329 pKa = 5.69VIARR333 pKa = 11.84KK334 pKa = 9.29PEE336 pKa = 3.9CCIMYY341 pKa = 10.3GLEE344 pKa = 4.27TIRR347 pKa = 11.84GSNQILDD354 pKa = 4.81KK355 pKa = 10.76IASDD359 pKa = 3.62YY360 pKa = 11.53KK361 pKa = 11.26SFFTIDD367 pKa = 2.65WSGFDD372 pKa = 4.51QRR374 pKa = 11.84LPWVIVKK381 pKa = 9.66LFFTEE386 pKa = 4.23YY387 pKa = 9.97IPRR390 pKa = 11.84LLVVNHH396 pKa = 6.92GYY398 pKa = 11.03APTYY402 pKa = 9.02EE403 pKa = 4.25YY404 pKa = 10.34PSYY407 pKa = 10.98PDD409 pKa = 3.35LTTNDD414 pKa = 3.23MVSRR418 pKa = 11.84LTNLLTFLATWYY430 pKa = 10.46FNMVFVTADD439 pKa = 2.87GFSYY443 pKa = 10.54VRR445 pKa = 11.84EE446 pKa = 4.19HH447 pKa = 6.82AGVPSGMLNTQFLDD461 pKa = 3.44SFGNLFLLIDD471 pKa = 3.82GLIEE475 pKa = 4.54FGSTDD480 pKa = 4.17AEE482 pKa = 4.06IDD484 pKa = 4.38DD485 pKa = 3.94ILLFIMGDD493 pKa = 3.62DD494 pKa = 3.56NSAFTTWSITHH505 pKa = 7.0LEE507 pKa = 4.04QFVSFFEE514 pKa = 4.66TYY516 pKa = 10.23ALSRR520 pKa = 11.84YY521 pKa = 9.99GMVLSKK527 pKa = 9.46TKK529 pKa = 10.69SIITTLRR536 pKa = 11.84HH537 pKa = 5.96KK538 pKa = 10.54IEE540 pKa = 4.17TLSYY544 pKa = 9.4QCNFGHH550 pKa = 7.22PRR552 pKa = 11.84RR553 pKa = 11.84PIGKK557 pKa = 9.07LVAQLCFPEE566 pKa = 4.83RR567 pKa = 11.84GPRR570 pKa = 11.84PKK572 pKa = 10.75YY573 pKa = 8.73MSARR577 pKa = 11.84AVGMAWASCGQDD589 pKa = 2.84KK590 pKa = 9.23TFHH593 pKa = 6.55DD594 pKa = 4.92FCRR597 pKa = 11.84DD598 pKa = 3.13VYY600 pKa = 11.3HH601 pKa = 6.92EE602 pKa = 4.75FNDD605 pKa = 4.1DD606 pKa = 3.93RR607 pKa = 11.84ADD609 pKa = 4.03LDD611 pKa = 3.62EE612 pKa = 4.4SAYY615 pKa = 11.34LHH617 pKa = 6.24IQSHH621 pKa = 6.05LPGYY625 pKa = 10.41LKK627 pKa = 9.66IDD629 pKa = 3.35EE630 pKa = 4.52SVRR633 pKa = 11.84QIVDD637 pKa = 3.74FQVFPSQQTVYY648 pKa = 9.09HH649 pKa = 5.79TVSRR653 pKa = 11.84WKK655 pKa = 10.9GPLSYY660 pKa = 10.33QPKK663 pKa = 8.42WDD665 pKa = 3.55LAHH668 pKa = 6.9FVNQPDD674 pKa = 3.99VVPPDD679 pKa = 3.4SVTLYY684 pKa = 10.05EE685 pKa = 4.5VWSEE689 pKa = 4.44LNFTPPILHH698 pKa = 6.6NLFF701 pKa = 4.17
Molecular weight: 81.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1338 |
637 |
701 |
669.0 |
76.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.904 ± 1.101 | 0.673 ± 0.242 |
6.203 ± 0.055 | 3.961 ± 0.661 |
6.054 ± 0.272 | 4.185 ± 0.142 |
3.737 ± 0.615 | 5.68 ± 0.231 |
4.111 ± 0.444 | 9.791 ± 0.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.018 ± 0.197 | 5.082 ± 1.126 |
6.876 ± 0.021 | 2.99 ± 0.747 |
4.933 ± 0.575 | 9.193 ± 1.213 |
7.25 ± 0.723 | 5.306 ± 0.021 |
1.046 ± 0.177 | 5.007 ± 0.519 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
