
Ilyobacter polytropus (strain ATCC 51220 / DSM 2926 / LMG 16218 / CuHBu1)
Taxonomy: cellular organisms; Bacteria; Fusobacteria; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Ilyobacter; Ilyobacter polytropus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
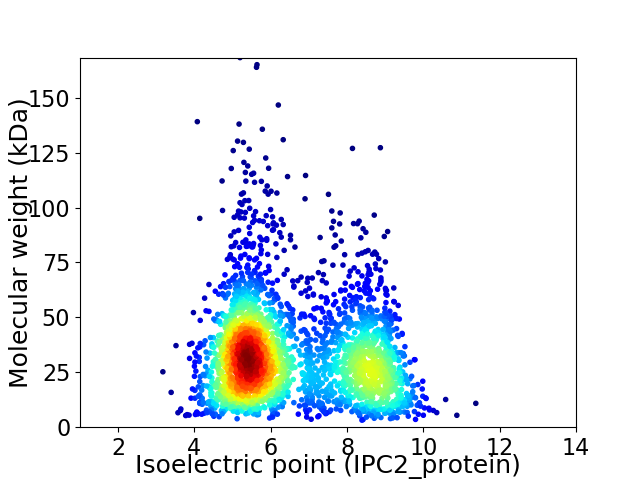
Virtual 2D-PAGE plot for 2859 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E3H6Z4|E3H6Z4_ILYPC Binding-protein-dependent transport systems inner membrane component OS=Ilyobacter polytropus (strain ATCC 51220 / DSM 2926 / LMG 16218 / CuHBu1) OX=572544 GN=Ilyop_0726 PE=3 SV=1
MM1 pKa = 7.0KK2 pKa = 10.14RR3 pKa = 11.84VMIFLMFLILVSGMAYY19 pKa = 10.08SEE21 pKa = 4.06TRR23 pKa = 11.84TVYY26 pKa = 10.95LIVPADD32 pKa = 3.91AGTLSYY38 pKa = 11.05YY39 pKa = 10.98AVDD42 pKa = 3.76EE43 pKa = 4.62YY44 pKa = 11.01EE45 pKa = 5.0AEE47 pKa = 4.29VTITASTEE55 pKa = 3.91TTSYY59 pKa = 10.91LGNSNLKK66 pKa = 10.4LVTLDD71 pKa = 4.03ASTSDD76 pKa = 3.49TFKK79 pKa = 11.42LMAKK83 pKa = 10.15YY84 pKa = 10.21KK85 pKa = 10.98LNDD88 pKa = 3.03KK89 pKa = 10.85YY90 pKa = 11.73YY91 pKa = 10.94FLISSNTDD99 pKa = 3.0TEE101 pKa = 4.45SDD103 pKa = 3.77DD104 pKa = 3.6NLEE107 pKa = 3.86ILEE110 pKa = 4.46TNFVSTNSPSEE121 pKa = 4.11VYY123 pKa = 10.67NVFGSFDD130 pKa = 4.45DD131 pKa = 3.99YY132 pKa = 11.52PGIKK136 pKa = 9.61IQDD139 pKa = 3.82DD140 pKa = 3.81VRR142 pKa = 11.84LSMIPVTDD150 pKa = 3.69EE151 pKa = 3.72FTEE154 pKa = 4.54FTDD157 pKa = 5.24GIVDD161 pKa = 3.8EE162 pKa = 5.58VYY164 pKa = 10.92LSDD167 pKa = 6.13DD168 pKa = 3.75DD169 pKa = 3.73EE170 pKa = 4.9TYY172 pKa = 11.77NNVAVNGYY180 pKa = 10.8AGISPNNSTTFTIKK194 pKa = 10.62EE195 pKa = 4.12KK196 pKa = 10.75DD197 pKa = 3.6SEE199 pKa = 4.6TEE201 pKa = 3.93TVYY204 pKa = 11.13TDD206 pKa = 4.14LKK208 pKa = 10.0TISDD212 pKa = 4.58PLTYY216 pKa = 10.36SYY218 pKa = 11.43TDD220 pKa = 3.72DD221 pKa = 5.48DD222 pKa = 5.51GNDD225 pKa = 3.43HH226 pKa = 6.08YY227 pKa = 10.12TVTVTYY233 pKa = 10.58AQFRR237 pKa = 11.84AKK239 pKa = 8.9NTNSSTFTYY248 pKa = 10.33DD249 pKa = 3.0GKK251 pKa = 10.72NDD253 pKa = 3.52GNEE256 pKa = 4.28TYY258 pKa = 10.12TLTGEE263 pKa = 4.32MVLDD267 pKa = 4.12LYY269 pKa = 11.12AYY271 pKa = 10.74NSGQGSYY278 pKa = 7.47WMEE281 pKa = 4.77IIPEE285 pKa = 4.36GDD287 pKa = 3.47TVVLIDD293 pKa = 5.43DD294 pKa = 3.99IGVTAEE300 pKa = 4.15RR301 pKa = 11.84EE302 pKa = 3.46IDD304 pKa = 3.6ATSKK308 pKa = 11.15ASDD311 pKa = 3.28KK312 pKa = 11.26SKK314 pKa = 11.13SRR316 pKa = 11.84DD317 pKa = 3.65TNIKK321 pKa = 9.44TSLIDD326 pKa = 3.31SDD328 pKa = 3.98NFIIKK333 pKa = 10.36NIEE336 pKa = 3.75EE337 pKa = 4.09
MM1 pKa = 7.0KK2 pKa = 10.14RR3 pKa = 11.84VMIFLMFLILVSGMAYY19 pKa = 10.08SEE21 pKa = 4.06TRR23 pKa = 11.84TVYY26 pKa = 10.95LIVPADD32 pKa = 3.91AGTLSYY38 pKa = 11.05YY39 pKa = 10.98AVDD42 pKa = 3.76EE43 pKa = 4.62YY44 pKa = 11.01EE45 pKa = 5.0AEE47 pKa = 4.29VTITASTEE55 pKa = 3.91TTSYY59 pKa = 10.91LGNSNLKK66 pKa = 10.4LVTLDD71 pKa = 4.03ASTSDD76 pKa = 3.49TFKK79 pKa = 11.42LMAKK83 pKa = 10.15YY84 pKa = 10.21KK85 pKa = 10.98LNDD88 pKa = 3.03KK89 pKa = 10.85YY90 pKa = 11.73YY91 pKa = 10.94FLISSNTDD99 pKa = 3.0TEE101 pKa = 4.45SDD103 pKa = 3.77DD104 pKa = 3.6NLEE107 pKa = 3.86ILEE110 pKa = 4.46TNFVSTNSPSEE121 pKa = 4.11VYY123 pKa = 10.67NVFGSFDD130 pKa = 4.45DD131 pKa = 3.99YY132 pKa = 11.52PGIKK136 pKa = 9.61IQDD139 pKa = 3.82DD140 pKa = 3.81VRR142 pKa = 11.84LSMIPVTDD150 pKa = 3.69EE151 pKa = 3.72FTEE154 pKa = 4.54FTDD157 pKa = 5.24GIVDD161 pKa = 3.8EE162 pKa = 5.58VYY164 pKa = 10.92LSDD167 pKa = 6.13DD168 pKa = 3.75DD169 pKa = 3.73EE170 pKa = 4.9TYY172 pKa = 11.77NNVAVNGYY180 pKa = 10.8AGISPNNSTTFTIKK194 pKa = 10.62EE195 pKa = 4.12KK196 pKa = 10.75DD197 pKa = 3.6SEE199 pKa = 4.6TEE201 pKa = 3.93TVYY204 pKa = 11.13TDD206 pKa = 4.14LKK208 pKa = 10.0TISDD212 pKa = 4.58PLTYY216 pKa = 10.36SYY218 pKa = 11.43TDD220 pKa = 3.72DD221 pKa = 5.48DD222 pKa = 5.51GNDD225 pKa = 3.43HH226 pKa = 6.08YY227 pKa = 10.12TVTVTYY233 pKa = 10.58AQFRR237 pKa = 11.84AKK239 pKa = 8.9NTNSSTFTYY248 pKa = 10.33DD249 pKa = 3.0GKK251 pKa = 10.72NDD253 pKa = 3.52GNEE256 pKa = 4.28TYY258 pKa = 10.12TLTGEE263 pKa = 4.32MVLDD267 pKa = 4.12LYY269 pKa = 11.12AYY271 pKa = 10.74NSGQGSYY278 pKa = 7.47WMEE281 pKa = 4.77IIPEE285 pKa = 4.36GDD287 pKa = 3.47TVVLIDD293 pKa = 5.43DD294 pKa = 3.99IGVTAEE300 pKa = 4.15RR301 pKa = 11.84EE302 pKa = 3.46IDD304 pKa = 3.6ATSKK308 pKa = 11.15ASDD311 pKa = 3.28KK312 pKa = 11.26SKK314 pKa = 11.13SRR316 pKa = 11.84DD317 pKa = 3.65TNIKK321 pKa = 9.44TSLIDD326 pKa = 3.31SDD328 pKa = 3.98NFIIKK333 pKa = 10.36NIEE336 pKa = 3.75EE337 pKa = 4.09
Molecular weight: 37.72 kDa
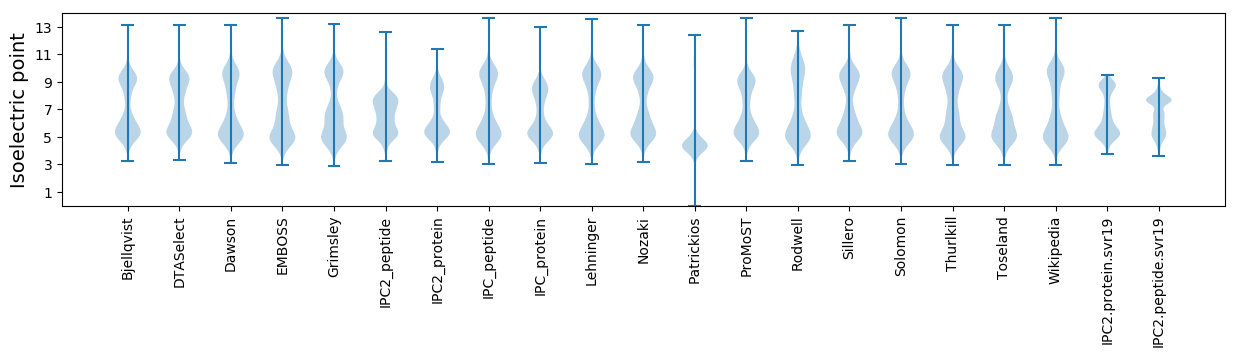
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3HBK3|E3HBK3_ILYPC Uncharacterized protein OS=Ilyobacter polytropus (strain ATCC 51220 / DSM 2926 / LMG 16218 / CuHBu1) OX=572544 GN=Ilyop_1928 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.3KK3 pKa = 10.24RR4 pKa = 11.84LVLLLTLILIVSSVSFAATSSRR26 pKa = 11.84RR27 pKa = 11.84ATSGGWAVSSRR38 pKa = 11.84RR39 pKa = 11.84ATSGGWAVSSRR50 pKa = 11.84RR51 pKa = 11.84ATSGGWAVSSRR62 pKa = 11.84RR63 pKa = 11.84ATSGGWAVSSRR74 pKa = 11.84RR75 pKa = 11.84ATSGGWAVSSRR86 pKa = 11.84RR87 pKa = 11.84ATSGGWAVSSRR98 pKa = 11.84RR99 pKa = 11.84ATSGGWW105 pKa = 2.96
MM1 pKa = 7.74KK2 pKa = 10.3KK3 pKa = 10.24RR4 pKa = 11.84LVLLLTLILIVSSVSFAATSSRR26 pKa = 11.84RR27 pKa = 11.84ATSGGWAVSSRR38 pKa = 11.84RR39 pKa = 11.84ATSGGWAVSSRR50 pKa = 11.84RR51 pKa = 11.84ATSGGWAVSSRR62 pKa = 11.84RR63 pKa = 11.84ATSGGWAVSSRR74 pKa = 11.84RR75 pKa = 11.84ATSGGWAVSSRR86 pKa = 11.84RR87 pKa = 11.84ATSGGWAVSSRR98 pKa = 11.84RR99 pKa = 11.84ATSGGWW105 pKa = 2.96
Molecular weight: 10.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
873442 |
30 |
1489 |
305.5 |
34.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.802 ± 0.052 | 0.943 ± 0.019 |
5.464 ± 0.038 | 8.167 ± 0.049 |
4.631 ± 0.037 | 7.092 ± 0.042 |
1.473 ± 0.017 | 9.146 ± 0.046 |
9.398 ± 0.051 | 9.104 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.822 ± 0.021 | 5.088 ± 0.032 |
2.923 ± 0.025 | 2.102 ± 0.018 |
3.656 ± 0.031 | 6.222 ± 0.035 |
4.79 ± 0.03 | 6.542 ± 0.035 |
0.703 ± 0.014 | 3.936 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
