
Pectobacterium phage DU_PP_II
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Unyawovirus; Pectobacterium virus DUPPII
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
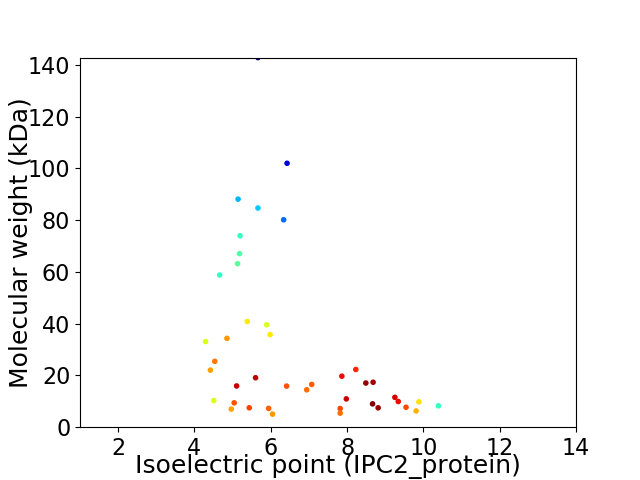
Virtual 2D-PAGE plot for 42 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
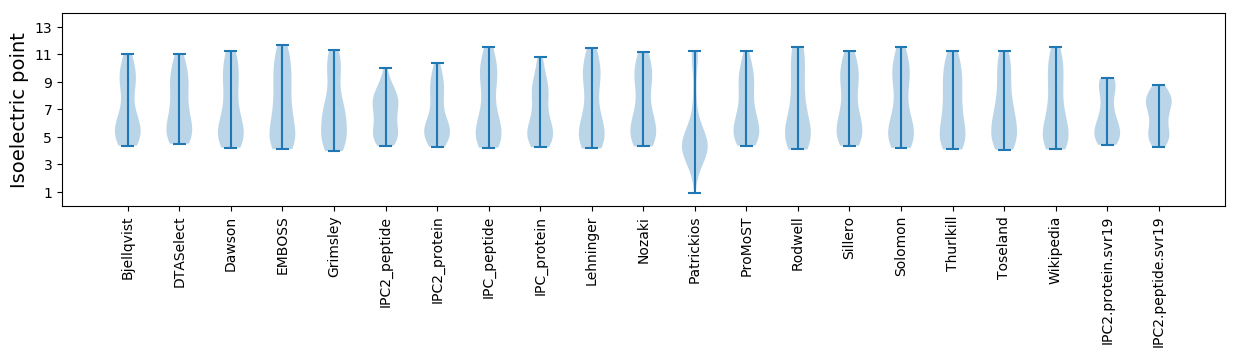
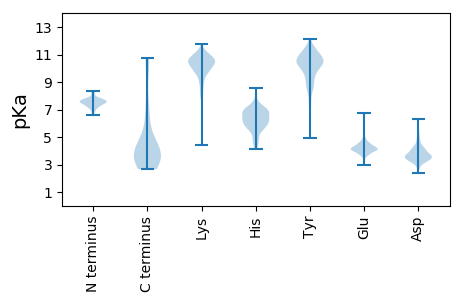
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2W5W2|A0A2D2W5W2_9CAUD Inhibitor of host bacterial RNA polymerase OS=Pectobacterium phage DU_PP_II OX=2041489 GN=P2B40kb_p010 PE=4 SV=1
MM1 pKa = 7.7AEE3 pKa = 4.14SNADD7 pKa = 3.26VYY9 pKa = 11.72ASFGVNSAVVTGGSQEE25 pKa = 3.99EE26 pKa = 4.58HH27 pKa = 5.48TQNMLALDD35 pKa = 3.7VATRR39 pKa = 11.84DD40 pKa = 3.47GDD42 pKa = 3.96DD43 pKa = 4.91LISITEE49 pKa = 4.11DD50 pKa = 3.54APVDD54 pKa = 4.27LYY56 pKa = 11.82ANSDD60 pKa = 3.3KK61 pKa = 10.73FANPEE66 pKa = 4.1DD67 pKa = 4.34DD68 pKa = 3.85NGFIQVRR75 pKa = 11.84IGDD78 pKa = 3.81GSDD81 pKa = 3.59PEE83 pKa = 4.52GQTTGQPEE91 pKa = 4.46VTTTDD96 pKa = 3.22DD97 pKa = 4.97VEE99 pKa = 4.29FQQLGEE105 pKa = 4.25IPTNLTDD112 pKa = 3.89TSQKK116 pKa = 10.64LADD119 pKa = 4.32HH120 pKa = 6.52EE121 pKa = 5.59AGFQTIVAQAQEE133 pKa = 4.28KK134 pKa = 9.88GVPEE138 pKa = 4.21DD139 pKa = 3.64SITRR143 pKa = 11.84IQSEE147 pKa = 4.38YY148 pKa = 10.24QGDD151 pKa = 4.37GISEE155 pKa = 3.68QSYY158 pKa = 11.25AEE160 pKa = 4.34LEE162 pKa = 3.93AAGYY166 pKa = 9.82SRR168 pKa = 11.84QFVDD172 pKa = 4.72SYY174 pKa = 11.59ISGQEE179 pKa = 3.97ALVDD183 pKa = 3.99AYY185 pKa = 11.19VNQVMDD191 pKa = 3.8FAGGRR196 pKa = 11.84EE197 pKa = 4.04AFQAVHH203 pKa = 5.72AHH205 pKa = 5.38MVATNPEE212 pKa = 3.86AAQTFEE218 pKa = 4.14TALGNRR224 pKa = 11.84DD225 pKa = 3.92MVTMKK230 pKa = 11.01ALLNLAGQSRR240 pKa = 11.84TKK242 pKa = 11.07AFGKK246 pKa = 9.05PAEE249 pKa = 4.18RR250 pKa = 11.84SMISKK255 pKa = 9.29GAPASPAKK263 pKa = 10.33PEE265 pKa = 4.01GKK267 pKa = 10.35AKK269 pKa = 10.58VEE271 pKa = 4.1PFASQQEE278 pKa = 4.58LIKK281 pKa = 11.12AMSDD285 pKa = 2.75DD286 pKa = 4.39RR287 pKa = 11.84YY288 pKa = 10.82RR289 pKa = 11.84SDD291 pKa = 2.79AAYY294 pKa = 10.0RR295 pKa = 11.84RR296 pKa = 11.84SVEE299 pKa = 3.73LRR301 pKa = 11.84VINSAFF307 pKa = 3.16
MM1 pKa = 7.7AEE3 pKa = 4.14SNADD7 pKa = 3.26VYY9 pKa = 11.72ASFGVNSAVVTGGSQEE25 pKa = 3.99EE26 pKa = 4.58HH27 pKa = 5.48TQNMLALDD35 pKa = 3.7VATRR39 pKa = 11.84DD40 pKa = 3.47GDD42 pKa = 3.96DD43 pKa = 4.91LISITEE49 pKa = 4.11DD50 pKa = 3.54APVDD54 pKa = 4.27LYY56 pKa = 11.82ANSDD60 pKa = 3.3KK61 pKa = 10.73FANPEE66 pKa = 4.1DD67 pKa = 4.34DD68 pKa = 3.85NGFIQVRR75 pKa = 11.84IGDD78 pKa = 3.81GSDD81 pKa = 3.59PEE83 pKa = 4.52GQTTGQPEE91 pKa = 4.46VTTTDD96 pKa = 3.22DD97 pKa = 4.97VEE99 pKa = 4.29FQQLGEE105 pKa = 4.25IPTNLTDD112 pKa = 3.89TSQKK116 pKa = 10.64LADD119 pKa = 4.32HH120 pKa = 6.52EE121 pKa = 5.59AGFQTIVAQAQEE133 pKa = 4.28KK134 pKa = 9.88GVPEE138 pKa = 4.21DD139 pKa = 3.64SITRR143 pKa = 11.84IQSEE147 pKa = 4.38YY148 pKa = 10.24QGDD151 pKa = 4.37GISEE155 pKa = 3.68QSYY158 pKa = 11.25AEE160 pKa = 4.34LEE162 pKa = 3.93AAGYY166 pKa = 9.82SRR168 pKa = 11.84QFVDD172 pKa = 4.72SYY174 pKa = 11.59ISGQEE179 pKa = 3.97ALVDD183 pKa = 3.99AYY185 pKa = 11.19VNQVMDD191 pKa = 3.8FAGGRR196 pKa = 11.84EE197 pKa = 4.04AFQAVHH203 pKa = 5.72AHH205 pKa = 5.38MVATNPEE212 pKa = 3.86AAQTFEE218 pKa = 4.14TALGNRR224 pKa = 11.84DD225 pKa = 3.92MVTMKK230 pKa = 11.01ALLNLAGQSRR240 pKa = 11.84TKK242 pKa = 11.07AFGKK246 pKa = 9.05PAEE249 pKa = 4.18RR250 pKa = 11.84SMISKK255 pKa = 9.29GAPASPAKK263 pKa = 10.33PEE265 pKa = 4.01GKK267 pKa = 10.35AKK269 pKa = 10.58VEE271 pKa = 4.1PFASQQEE278 pKa = 4.58LIKK281 pKa = 11.12AMSDD285 pKa = 2.75DD286 pKa = 4.39RR287 pKa = 11.84YY288 pKa = 10.82RR289 pKa = 11.84SDD291 pKa = 2.79AAYY294 pKa = 10.0RR295 pKa = 11.84RR296 pKa = 11.84SVEE299 pKa = 3.73LRR301 pKa = 11.84VINSAFF307 pKa = 3.16
Molecular weight: 33.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2W5U2|A0A2D2W5U2_9CAUD Serine recombinase OS=Pectobacterium phage DU_PP_II OX=2041489 GN=P2B40kb_p001 PE=4 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84LHH4 pKa = 6.56YY5 pKa = 10.32NSSNGIFSVRR15 pKa = 11.84RR16 pKa = 11.84PDD18 pKa = 4.19RR19 pKa = 11.84STQSASGKK27 pKa = 8.08HH28 pKa = 4.78QKK30 pKa = 10.6LPIIGDD36 pKa = 3.62IVEE39 pKa = 4.7LSPKK43 pKa = 10.02VFLLITRR50 pKa = 11.84GEE52 pKa = 4.09FVEE55 pKa = 4.38ATKK58 pKa = 9.45GTRR61 pKa = 11.84PFMQAVVTSWPKK73 pKa = 8.53TRR75 pKa = 11.84LVIKK79 pKa = 10.13RR80 pKa = 11.84IKK82 pKa = 10.22EE83 pKa = 3.78IIKK86 pKa = 10.44
MM1 pKa = 7.77RR2 pKa = 11.84LHH4 pKa = 6.56YY5 pKa = 10.32NSSNGIFSVRR15 pKa = 11.84RR16 pKa = 11.84PDD18 pKa = 4.19RR19 pKa = 11.84STQSASGKK27 pKa = 8.08HH28 pKa = 4.78QKK30 pKa = 10.6LPIIGDD36 pKa = 3.62IVEE39 pKa = 4.7LSPKK43 pKa = 10.02VFLLITRR50 pKa = 11.84GEE52 pKa = 4.09FVEE55 pKa = 4.38ATKK58 pKa = 9.45GTRR61 pKa = 11.84PFMQAVVTSWPKK73 pKa = 8.53TRR75 pKa = 11.84LVIKK79 pKa = 10.13RR80 pKa = 11.84IKK82 pKa = 10.22EE83 pKa = 3.78IIKK86 pKa = 10.44
Molecular weight: 9.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11648 |
48 |
1328 |
277.3 |
30.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.989 ± 0.522 | 0.91 ± 0.172 |
6.053 ± 0.179 | 6.533 ± 0.318 |
3.735 ± 0.166 | 7.95 ± 0.287 |
1.743 ± 0.176 | 5.263 ± 0.142 |
6.156 ± 0.387 | 8.079 ± 0.281 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.618 ± 0.174 | 4.507 ± 0.295 |
3.872 ± 0.228 | 4.301 ± 0.319 |
5.237 ± 0.181 | 6.585 ± 0.256 |
5.958 ± 0.285 | 6.739 ± 0.311 |
1.52 ± 0.164 | 3.254 ± 0.197 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
