
Polaribacter sp. ALD11
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter; unclassified Polaribacter
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
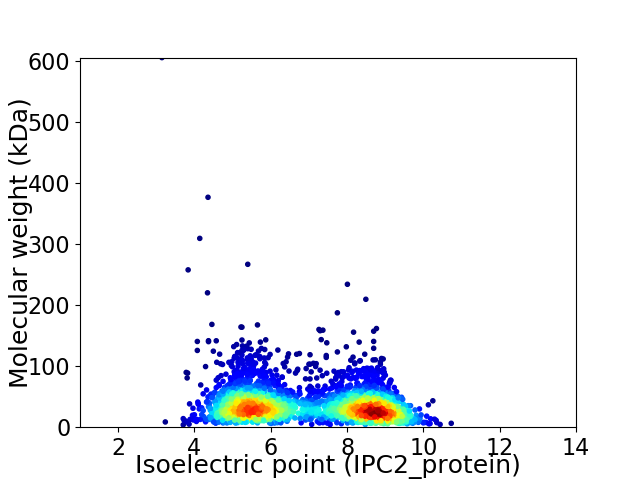
Virtual 2D-PAGE plot for 2976 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8XMY1|A0A2K8XMY1_9FLAO Uncharacterized protein OS=Polaribacter sp. ALD11 OX=2058137 GN=CW731_02915 PE=4 SV=1
MM1 pKa = 7.75SYY3 pKa = 10.41QAVVRR8 pKa = 11.84DD9 pKa = 3.75ASNTLVSTQAVGMQISILQGSTPVYY34 pKa = 10.86VEE36 pKa = 4.06AQTPTTNANGLVSLEE51 pKa = 4.2IGTGTLLSGDD61 pKa = 3.6FTTINWANDD70 pKa = 2.81AYY72 pKa = 10.63FIKK75 pKa = 10.09TEE77 pKa = 3.98TDD79 pKa = 3.06PTGGTNYY86 pKa = 9.61TITVTTQLMSVPYY99 pKa = 9.97ALHH102 pKa = 6.88AKK104 pKa = 7.68TAEE107 pKa = 4.52SITGTINYY115 pKa = 8.01TEE117 pKa = 4.12TDD119 pKa = 3.72SVFRR123 pKa = 11.84ASIANTITAIDD134 pKa = 3.51TTNWNNHH141 pKa = 4.45TVDD144 pKa = 3.64TDD146 pKa = 3.34THH148 pKa = 6.72IDD150 pKa = 3.48STGVANLGFNTGAIITEE167 pKa = 3.91TDD169 pKa = 3.92PIFRR173 pKa = 11.84ASIANGITAIDD184 pKa = 3.44TANWNNHH191 pKa = 4.31TVDD194 pKa = 3.64TDD196 pKa = 3.34THH198 pKa = 6.72IDD200 pKa = 3.48STGVANLGFNAGGIITEE217 pKa = 4.08TDD219 pKa = 3.31PVFRR223 pKa = 11.84ASITNGITAIDD234 pKa = 3.64TVNWNNKK241 pKa = 9.13LSTEE245 pKa = 3.85IDD247 pKa = 3.36GSVTNEE253 pKa = 3.78IQVLTISNDD262 pKa = 3.3TLFLSKK268 pKa = 10.85GGLVKK273 pKa = 10.66LPAAAVVFSGNYY285 pKa = 10.04ADD287 pKa = 4.5LTNIPAYY294 pKa = 9.85IADD297 pKa = 4.32GDD299 pKa = 5.1DD300 pKa = 3.32NTQLNEE306 pKa = 4.11TQVDD310 pKa = 3.57AFVANNGYY318 pKa = 8.25LTTEE322 pKa = 3.75VDD324 pKa = 3.58GSVTNEE330 pKa = 3.51IEE332 pKa = 4.27LPTGGTNGQVLKK344 pKa = 10.17TDD346 pKa = 3.32GGGNYY351 pKa = 10.22AWVNQTTDD359 pKa = 2.97TDD361 pKa = 3.92TNTQLNEE368 pKa = 4.07TQVDD372 pKa = 3.57AFVANNGYY380 pKa = 8.25LTTEE384 pKa = 3.75VDD386 pKa = 3.58GSVTNEE392 pKa = 3.51IEE394 pKa = 4.27LPTGGTNGQVLKK406 pKa = 10.17TDD408 pKa = 3.32GGGNYY413 pKa = 10.22AWVNQTTDD421 pKa = 2.97TDD423 pKa = 3.92TNTQLNEE430 pKa = 4.07TQVDD434 pKa = 3.57AFVANNGYY442 pKa = 8.25LTTEE446 pKa = 3.75VDD448 pKa = 3.58GSVTNEE454 pKa = 3.51IEE456 pKa = 4.27LPTGGTNGQVLKK468 pKa = 10.17TDD470 pKa = 3.58GSGNYY475 pKa = 9.82AWVNQTTDD483 pKa = 3.11TDD485 pKa = 3.3TDD487 pKa = 3.69TQLNEE492 pKa = 4.11TQVDD496 pKa = 3.57AFVANNGYY504 pKa = 8.25LTTEE508 pKa = 3.75VDD510 pKa = 3.58GSVTNEE516 pKa = 3.51IEE518 pKa = 4.27LPTGGTNGQVLKK530 pKa = 10.17TDD532 pKa = 3.58GSGNYY537 pKa = 9.82AWVNQTTDD545 pKa = 3.01TDD547 pKa = 3.8TDD549 pKa = 3.49THH551 pKa = 7.07IDD553 pKa = 3.36STGIANLGYY562 pKa = 9.89IPGVHH567 pKa = 5.56TVNTDD572 pKa = 2.88AQTLKK577 pKa = 10.8ASQTGDD583 pKa = 3.04TLTINGGNSVLIPGISLFNYY603 pKa = 9.32QPQIQIGEE611 pKa = 4.43FYY613 pKa = 10.87AGGVIFWLDD622 pKa = 3.45GNGGGLVVSIVDD634 pKa = 3.37QSTRR638 pKa = 11.84AQWGCHH644 pKa = 3.95GTHH647 pKa = 7.16LAGAEE652 pKa = 4.16GKK654 pKa = 10.49AIGTGAQNTADD665 pKa = 3.65IEE667 pKa = 4.53AGCTRR672 pKa = 11.84YY673 pKa = 8.4ATAADD678 pKa = 3.98LCANLTLNGYY688 pKa = 9.41NDD690 pKa = 3.65WFLPSIDD697 pKa = 3.85EE698 pKa = 4.2LQAIFDD704 pKa = 3.61NRR706 pKa = 11.84AAINATAISNLGTSLGSYY724 pKa = 8.46YY725 pKa = 9.98WSSTEE730 pKa = 5.15DD731 pKa = 3.32YY732 pKa = 10.78NYY734 pKa = 9.54WALLYY739 pKa = 10.09HH740 pKa = 6.34FRR742 pKa = 11.84AGSVSEE748 pKa = 4.95DD749 pKa = 3.09KK750 pKa = 11.06FEE752 pKa = 4.11KK753 pKa = 10.34HH754 pKa = 5.11GVRR757 pKa = 11.84AVRR760 pKa = 11.84AFF762 pKa = 3.27
MM1 pKa = 7.75SYY3 pKa = 10.41QAVVRR8 pKa = 11.84DD9 pKa = 3.75ASNTLVSTQAVGMQISILQGSTPVYY34 pKa = 10.86VEE36 pKa = 4.06AQTPTTNANGLVSLEE51 pKa = 4.2IGTGTLLSGDD61 pKa = 3.6FTTINWANDD70 pKa = 2.81AYY72 pKa = 10.63FIKK75 pKa = 10.09TEE77 pKa = 3.98TDD79 pKa = 3.06PTGGTNYY86 pKa = 9.61TITVTTQLMSVPYY99 pKa = 9.97ALHH102 pKa = 6.88AKK104 pKa = 7.68TAEE107 pKa = 4.52SITGTINYY115 pKa = 8.01TEE117 pKa = 4.12TDD119 pKa = 3.72SVFRR123 pKa = 11.84ASIANTITAIDD134 pKa = 3.51TTNWNNHH141 pKa = 4.45TVDD144 pKa = 3.64TDD146 pKa = 3.34THH148 pKa = 6.72IDD150 pKa = 3.48STGVANLGFNTGAIITEE167 pKa = 3.91TDD169 pKa = 3.92PIFRR173 pKa = 11.84ASIANGITAIDD184 pKa = 3.44TANWNNHH191 pKa = 4.31TVDD194 pKa = 3.64TDD196 pKa = 3.34THH198 pKa = 6.72IDD200 pKa = 3.48STGVANLGFNAGGIITEE217 pKa = 4.08TDD219 pKa = 3.31PVFRR223 pKa = 11.84ASITNGITAIDD234 pKa = 3.64TVNWNNKK241 pKa = 9.13LSTEE245 pKa = 3.85IDD247 pKa = 3.36GSVTNEE253 pKa = 3.78IQVLTISNDD262 pKa = 3.3TLFLSKK268 pKa = 10.85GGLVKK273 pKa = 10.66LPAAAVVFSGNYY285 pKa = 10.04ADD287 pKa = 4.5LTNIPAYY294 pKa = 9.85IADD297 pKa = 4.32GDD299 pKa = 5.1DD300 pKa = 3.32NTQLNEE306 pKa = 4.11TQVDD310 pKa = 3.57AFVANNGYY318 pKa = 8.25LTTEE322 pKa = 3.75VDD324 pKa = 3.58GSVTNEE330 pKa = 3.51IEE332 pKa = 4.27LPTGGTNGQVLKK344 pKa = 10.17TDD346 pKa = 3.32GGGNYY351 pKa = 10.22AWVNQTTDD359 pKa = 2.97TDD361 pKa = 3.92TNTQLNEE368 pKa = 4.07TQVDD372 pKa = 3.57AFVANNGYY380 pKa = 8.25LTTEE384 pKa = 3.75VDD386 pKa = 3.58GSVTNEE392 pKa = 3.51IEE394 pKa = 4.27LPTGGTNGQVLKK406 pKa = 10.17TDD408 pKa = 3.32GGGNYY413 pKa = 10.22AWVNQTTDD421 pKa = 2.97TDD423 pKa = 3.92TNTQLNEE430 pKa = 4.07TQVDD434 pKa = 3.57AFVANNGYY442 pKa = 8.25LTTEE446 pKa = 3.75VDD448 pKa = 3.58GSVTNEE454 pKa = 3.51IEE456 pKa = 4.27LPTGGTNGQVLKK468 pKa = 10.17TDD470 pKa = 3.58GSGNYY475 pKa = 9.82AWVNQTTDD483 pKa = 3.11TDD485 pKa = 3.3TDD487 pKa = 3.69TQLNEE492 pKa = 4.11TQVDD496 pKa = 3.57AFVANNGYY504 pKa = 8.25LTTEE508 pKa = 3.75VDD510 pKa = 3.58GSVTNEE516 pKa = 3.51IEE518 pKa = 4.27LPTGGTNGQVLKK530 pKa = 10.17TDD532 pKa = 3.58GSGNYY537 pKa = 9.82AWVNQTTDD545 pKa = 3.01TDD547 pKa = 3.8TDD549 pKa = 3.49THH551 pKa = 7.07IDD553 pKa = 3.36STGIANLGYY562 pKa = 9.89IPGVHH567 pKa = 5.56TVNTDD572 pKa = 2.88AQTLKK577 pKa = 10.8ASQTGDD583 pKa = 3.04TLTINGGNSVLIPGISLFNYY603 pKa = 9.32QPQIQIGEE611 pKa = 4.43FYY613 pKa = 10.87AGGVIFWLDD622 pKa = 3.45GNGGGLVVSIVDD634 pKa = 3.37QSTRR638 pKa = 11.84AQWGCHH644 pKa = 3.95GTHH647 pKa = 7.16LAGAEE652 pKa = 4.16GKK654 pKa = 10.49AIGTGAQNTADD665 pKa = 3.65IEE667 pKa = 4.53AGCTRR672 pKa = 11.84YY673 pKa = 8.4ATAADD678 pKa = 3.98LCANLTLNGYY688 pKa = 9.41NDD690 pKa = 3.65WFLPSIDD697 pKa = 3.85EE698 pKa = 4.2LQAIFDD704 pKa = 3.61NRR706 pKa = 11.84AAINATAISNLGTSLGSYY724 pKa = 8.46YY725 pKa = 9.98WSSTEE730 pKa = 5.15DD731 pKa = 3.32YY732 pKa = 10.78NYY734 pKa = 9.54WALLYY739 pKa = 10.09HH740 pKa = 6.34FRR742 pKa = 11.84AGSVSEE748 pKa = 4.95DD749 pKa = 3.09KK750 pKa = 11.06FEE752 pKa = 4.11KK753 pKa = 10.34HH754 pKa = 5.11GVRR757 pKa = 11.84AVRR760 pKa = 11.84AFF762 pKa = 3.27
Molecular weight: 80.65 kDa
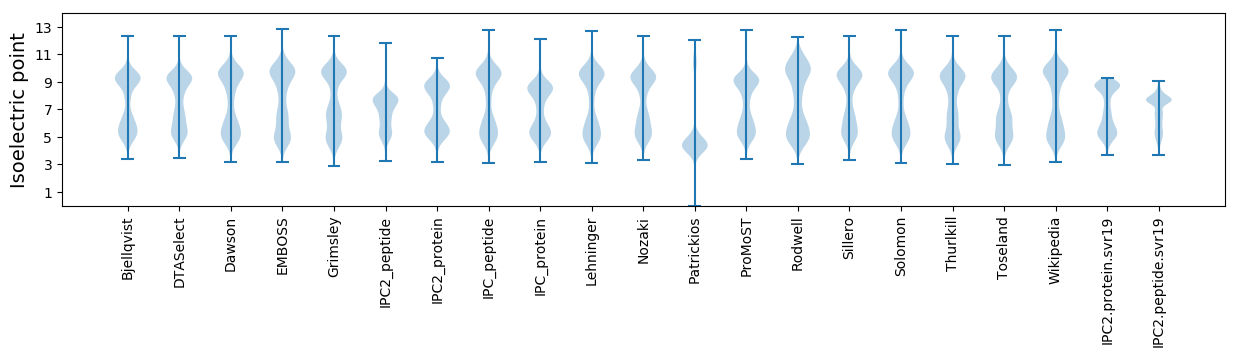
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8XU82|A0A2K8XU82_9FLAO Uncharacterized protein OS=Polaribacter sp. ALD11 OX=2058137 GN=CW731_15010 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 3.92TITTDD27 pKa = 3.73KK28 pKa = 10.91PEE30 pKa = 4.37KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.38RR39 pKa = 11.84SGGRR43 pKa = 11.84NNQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.58KK59 pKa = 9.49QKK61 pKa = 10.8YY62 pKa = 9.58RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.83RR69 pKa = 11.84DD70 pKa = 3.38KK71 pKa = 10.89QGIPATVKK79 pKa = 10.01TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALLSYY96 pKa = 11.49ADD98 pKa = 3.5GEE100 pKa = 4.22KK101 pKa = 10.25RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 9.81VGQVIVSGEE120 pKa = 4.23GIKK123 pKa = 10.52PEE125 pKa = 4.26IGNGMLLSEE134 pKa = 4.93IPLGTTISCIEE145 pKa = 4.03LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGSFAQLLAKK167 pKa = 9.53EE168 pKa = 4.49SKK170 pKa = 9.68YY171 pKa = 10.48ATVKK175 pKa = 10.6LPSGEE180 pKa = 3.84TRR182 pKa = 11.84YY183 pKa = 10.28ILLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.52ASNKK266 pKa = 9.94YY267 pKa = 8.99ILEE270 pKa = 4.09RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.2KK274 pKa = 9.94
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 3.92TITTDD27 pKa = 3.73KK28 pKa = 10.91PEE30 pKa = 4.37KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.38RR39 pKa = 11.84SGGRR43 pKa = 11.84NNQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.58KK59 pKa = 9.49QKK61 pKa = 10.8YY62 pKa = 9.58RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.83RR69 pKa = 11.84DD70 pKa = 3.38KK71 pKa = 10.89QGIPATVKK79 pKa = 10.01TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALLSYY96 pKa = 11.49ADD98 pKa = 3.5GEE100 pKa = 4.22KK101 pKa = 10.25RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 9.81VGQVIVSGEE120 pKa = 4.23GIKK123 pKa = 10.52PEE125 pKa = 4.26IGNGMLLSEE134 pKa = 4.93IPLGTTISCIEE145 pKa = 4.03LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGSFAQLLAKK167 pKa = 9.53EE168 pKa = 4.49SKK170 pKa = 9.68YY171 pKa = 10.48ATVKK175 pKa = 10.6LPSGEE180 pKa = 3.84TRR182 pKa = 11.84YY183 pKa = 10.28ILLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.52ASNKK266 pKa = 9.94YY267 pKa = 8.99ILEE270 pKa = 4.09RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.2KK274 pKa = 9.94
Molecular weight: 29.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1034009 |
36 |
5843 |
347.4 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.126 ± 0.042 | 0.662 ± 0.013 |
5.31 ± 0.047 | 6.636 ± 0.047 |
5.505 ± 0.045 | 6.168 ± 0.047 |
1.612 ± 0.022 | 8.42 ± 0.043 |
8.798 ± 0.061 | 9.216 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.0 ± 0.022 | 6.645 ± 0.053 |
3.065 ± 0.024 | 3.061 ± 0.023 |
3.139 ± 0.03 | 6.61 ± 0.043 |
6.005 ± 0.063 | 6.126 ± 0.033 |
0.974 ± 0.013 | 3.922 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
