
Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Nannocystineae; Kofleriaceae; Haliangium; Haliangium ochraceum
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
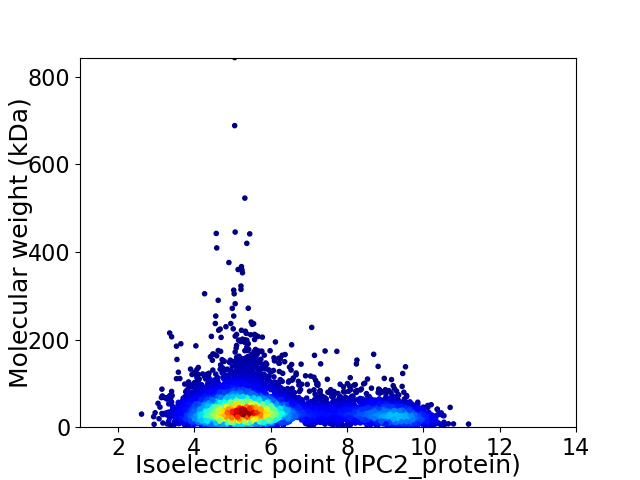
Virtual 2D-PAGE plot for 6684 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
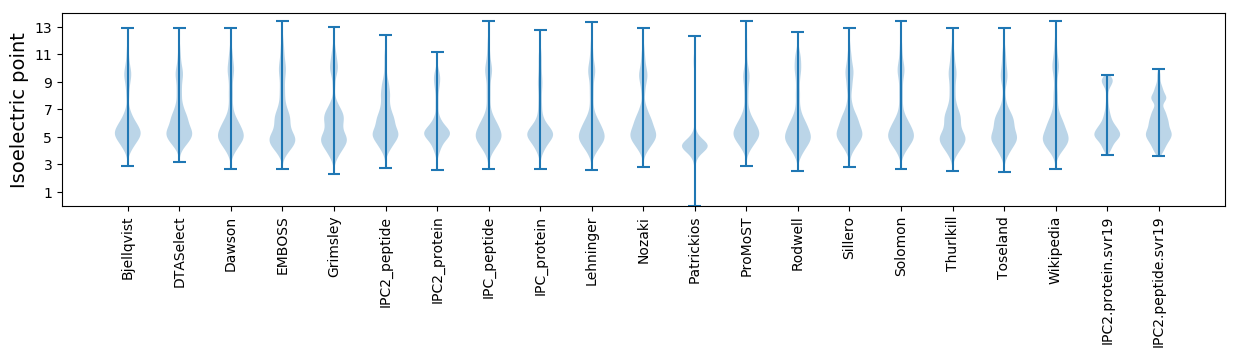
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0LXL5|D0LXL5_HALO1 Small ribosomal subunit biogenesis GTPase RsgA OS=Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) OX=502025 GN=rsgA PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 10.31RR3 pKa = 11.84IFSTTLTLCSLSLLLGACGGDD24 pKa = 3.54DD25 pKa = 3.46EE26 pKa = 6.06LSNEE30 pKa = 4.66DD31 pKa = 3.67YY32 pKa = 11.49DD33 pKa = 5.56DD34 pKa = 3.69IAVGVGSLVAVPGGEE49 pKa = 4.07GGEE52 pKa = 4.16TGSLNDD58 pKa = 3.51VLEE61 pKa = 4.44ASISAGTSAAEE72 pKa = 4.2GASSVEE78 pKa = 4.25IVVHH82 pKa = 6.41AGVSYY87 pKa = 9.83EE88 pKa = 3.88YY89 pKa = 11.06RR90 pKa = 11.84LDD92 pKa = 4.08CYY94 pKa = 10.84DD95 pKa = 4.16AAGALQDD102 pKa = 3.84ACGADD107 pKa = 3.54TDD109 pKa = 4.22SAEE112 pKa = 5.17ASVDD116 pKa = 2.95WSGEE120 pKa = 3.87LDD122 pKa = 3.73TPSYY126 pKa = 10.17DD127 pKa = 3.32ASIMRR132 pKa = 11.84VGDD135 pKa = 3.35WSLSGLQSDD144 pKa = 3.92TAVFSGTGSFDD155 pKa = 3.25IEE157 pKa = 4.51TSFSAMYY164 pKa = 10.27RR165 pKa = 11.84DD166 pKa = 3.86VEE168 pKa = 4.42RR169 pKa = 11.84TLSLSYY175 pKa = 10.47DD176 pKa = 3.24ASYY179 pKa = 11.83DD180 pKa = 4.48DD181 pKa = 3.54ITIDD185 pKa = 2.97IASRR189 pKa = 11.84RR190 pKa = 11.84VTGGAIHH197 pKa = 6.17YY198 pKa = 9.2NIDD201 pKa = 3.05GARR204 pKa = 11.84YY205 pKa = 9.02DD206 pKa = 3.57EE207 pKa = 4.55RR208 pKa = 11.84GARR211 pKa = 11.84TKK213 pKa = 10.36EE214 pKa = 3.41IEE216 pKa = 4.21FAVTADD222 pKa = 4.26LSFTGDD228 pKa = 3.37GEE230 pKa = 4.38ALLVLDD236 pKa = 4.65GSRR239 pKa = 11.84SYY241 pKa = 11.47VVDD244 pKa = 4.51LDD246 pKa = 3.92TGAVAAEE253 pKa = 4.06
MM1 pKa = 7.28KK2 pKa = 10.31RR3 pKa = 11.84IFSTTLTLCSLSLLLGACGGDD24 pKa = 3.54DD25 pKa = 3.46EE26 pKa = 6.06LSNEE30 pKa = 4.66DD31 pKa = 3.67YY32 pKa = 11.49DD33 pKa = 5.56DD34 pKa = 3.69IAVGVGSLVAVPGGEE49 pKa = 4.07GGEE52 pKa = 4.16TGSLNDD58 pKa = 3.51VLEE61 pKa = 4.44ASISAGTSAAEE72 pKa = 4.2GASSVEE78 pKa = 4.25IVVHH82 pKa = 6.41AGVSYY87 pKa = 9.83EE88 pKa = 3.88YY89 pKa = 11.06RR90 pKa = 11.84LDD92 pKa = 4.08CYY94 pKa = 10.84DD95 pKa = 4.16AAGALQDD102 pKa = 3.84ACGADD107 pKa = 3.54TDD109 pKa = 4.22SAEE112 pKa = 5.17ASVDD116 pKa = 2.95WSGEE120 pKa = 3.87LDD122 pKa = 3.73TPSYY126 pKa = 10.17DD127 pKa = 3.32ASIMRR132 pKa = 11.84VGDD135 pKa = 3.35WSLSGLQSDD144 pKa = 3.92TAVFSGTGSFDD155 pKa = 3.25IEE157 pKa = 4.51TSFSAMYY164 pKa = 10.27RR165 pKa = 11.84DD166 pKa = 3.86VEE168 pKa = 4.42RR169 pKa = 11.84TLSLSYY175 pKa = 10.47DD176 pKa = 3.24ASYY179 pKa = 11.83DD180 pKa = 4.48DD181 pKa = 3.54ITIDD185 pKa = 2.97IASRR189 pKa = 11.84RR190 pKa = 11.84VTGGAIHH197 pKa = 6.17YY198 pKa = 9.2NIDD201 pKa = 3.05GARR204 pKa = 11.84YY205 pKa = 9.02DD206 pKa = 3.57EE207 pKa = 4.55RR208 pKa = 11.84GARR211 pKa = 11.84TKK213 pKa = 10.36EE214 pKa = 3.41IEE216 pKa = 4.21FAVTADD222 pKa = 4.26LSFTGDD228 pKa = 3.37GEE230 pKa = 4.38ALLVLDD236 pKa = 4.65GSRR239 pKa = 11.84SYY241 pKa = 11.47VVDD244 pKa = 4.51LDD246 pKa = 3.92TGAVAAEE253 pKa = 4.06
Molecular weight: 26.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0LUI2|D0LUI2_HALO1 SsrA-binding protein OS=Haliangium ochraceum (strain DSM 14365 / JCM 11303 / SMP-2) OX=502025 GN=smpB PE=3 SV=1
MM1 pKa = 7.01ATGRR5 pKa = 11.84RR6 pKa = 11.84TKK8 pKa = 10.47RR9 pKa = 11.84SLKK12 pKa = 10.16DD13 pKa = 3.49RR14 pKa = 11.84PKK16 pKa = 10.44HH17 pKa = 4.72RR18 pKa = 11.84QARR21 pKa = 11.84ALKK24 pKa = 8.4TRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.77AKK31 pKa = 9.36QAASRR36 pKa = 11.84KK37 pKa = 9.2RR38 pKa = 11.84GLNKK42 pKa = 9.73SRR44 pKa = 11.84ARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84TRR50 pKa = 11.84QGRR53 pKa = 11.84SGKK56 pKa = 10.07RR57 pKa = 11.84VIRR60 pKa = 4.17
MM1 pKa = 7.01ATGRR5 pKa = 11.84RR6 pKa = 11.84TKK8 pKa = 10.47RR9 pKa = 11.84SLKK12 pKa = 10.16DD13 pKa = 3.49RR14 pKa = 11.84PKK16 pKa = 10.44HH17 pKa = 4.72RR18 pKa = 11.84QARR21 pKa = 11.84ALKK24 pKa = 8.4TRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.77AKK31 pKa = 9.36QAASRR36 pKa = 11.84KK37 pKa = 9.2RR38 pKa = 11.84GLNKK42 pKa = 9.73SRR44 pKa = 11.84ARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84TRR50 pKa = 11.84QGRR53 pKa = 11.84SGKK56 pKa = 10.07RR57 pKa = 11.84VIRR60 pKa = 4.17
Molecular weight: 7.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2756058 |
32 |
7712 |
412.3 |
44.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.572 ± 0.056 | 1.148 ± 0.02 |
6.324 ± 0.03 | 6.704 ± 0.023 |
3.115 ± 0.017 | 8.563 ± 0.032 |
2.234 ± 0.014 | 3.714 ± 0.022 |
2.101 ± 0.026 | 10.364 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.87 ± 0.012 | 2.002 ± 0.019 |
5.547 ± 0.022 | 3.227 ± 0.018 |
8.336 ± 0.035 | 5.843 ± 0.021 |
4.613 ± 0.019 | 7.244 ± 0.026 |
1.277 ± 0.012 | 2.203 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
