
Caldilinea aerophila (strain DSM 14535 / JCM 11387 / NBRC 104270 / STL-6-O1)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Caldilineae; Caldilineales; Caldilineaceae; Caldilinea; Caldilinea aerophila
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
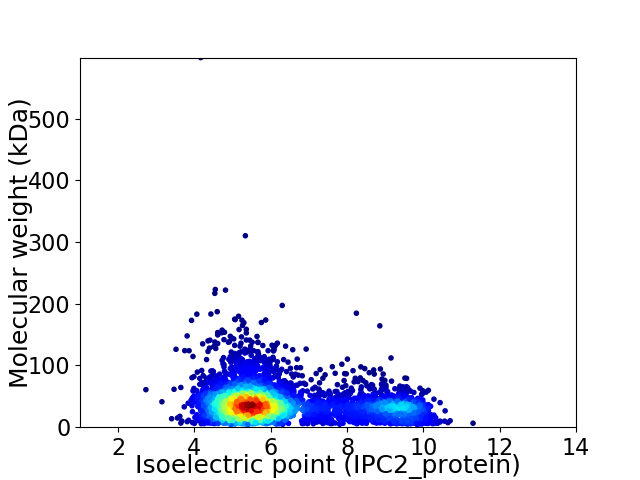
Virtual 2D-PAGE plot for 4097 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0I1K4|I0I1K4_CALAS Putative oligopeptide ABC transporter permease protein OS=Caldilinea aerophila (strain DSM 14535 / JCM 11387 / NBRC 104270 / STL-6-O1) OX=926550 GN=CLDAP_11020 PE=3 SV=1
MM1 pKa = 7.49LALALSALPTPLLAAPFQQEE21 pKa = 4.4VPVSDD26 pKa = 3.86GLTLEE31 pKa = 4.69ALANLTYY38 pKa = 10.37RR39 pKa = 11.84SEE41 pKa = 4.14LTPSGEE47 pKa = 3.74ITLVDD52 pKa = 3.94GRR54 pKa = 11.84YY55 pKa = 8.91EE56 pKa = 4.15DD57 pKa = 3.47PANRR61 pKa = 11.84VFAVLAAEE69 pKa = 4.25PMAFGTINGQDD80 pKa = 3.18AAAVLLAEE88 pKa = 4.33SSGAGSVFITLAIVVDD104 pKa = 3.99QDD106 pKa = 3.86GEE108 pKa = 4.42PVNVASTLLGDD119 pKa = 3.89RR120 pKa = 11.84VDD122 pKa = 4.17VYY124 pKa = 11.56ALEE127 pKa = 4.85IDD129 pKa = 4.19GSRR132 pKa = 11.84ILVDD136 pKa = 3.51MVRR139 pKa = 11.84QGPNDD144 pKa = 3.66PMCCPTEE151 pKa = 4.17VVRR154 pKa = 11.84QIYY157 pKa = 10.6ALGEE161 pKa = 4.03EE162 pKa = 4.55GLILAGVNLIGSGVQAALNVSPEE185 pKa = 3.72GGYY188 pKa = 7.15QTKK191 pKa = 10.2VMPATPYY198 pKa = 10.46AISALIPTGAPIHH211 pKa = 5.87TLLTFGDD218 pKa = 4.67DD219 pKa = 3.69DD220 pKa = 4.82PEE222 pKa = 4.56TVFLNGGPYY231 pKa = 8.91IAIYY235 pKa = 9.26PVQEE239 pKa = 4.46FVRR242 pKa = 11.84LWEE245 pKa = 4.01EE246 pKa = 4.25AGDD249 pKa = 3.72LTIANLVEE257 pKa = 3.93EE258 pKa = 5.15LKK260 pKa = 10.8RR261 pKa = 11.84ILTEE265 pKa = 4.09QPASLAAPLPIVPPADD281 pKa = 3.25VDD283 pKa = 4.17DD284 pKa = 5.35LVAQVVYY291 pKa = 11.05LEE293 pKa = 5.18GEE295 pKa = 4.24DD296 pKa = 3.63FTGVRR301 pKa = 11.84FIARR305 pKa = 11.84SVRR308 pKa = 11.84NGIVSADD315 pKa = 3.41GQLAYY320 pKa = 10.49HH321 pKa = 5.78FTGLTDD327 pKa = 3.72DD328 pKa = 3.5SRR330 pKa = 11.84YY331 pKa = 10.44LIAAQWPVEE340 pKa = 4.19TTAALTALDD349 pKa = 4.39SAPDD353 pKa = 4.02SAWTPALSSLDD364 pKa = 3.61VLIEE368 pKa = 3.97SLTIQGPEE376 pKa = 3.55WVTVEE381 pKa = 3.92EE382 pKa = 4.55LANLTYY388 pKa = 10.82DD389 pKa = 3.27SVMLEE394 pKa = 4.12KK395 pKa = 10.33PVTLSSGVYY404 pKa = 8.63TEE406 pKa = 4.29TVIPDD411 pKa = 3.53VPAGVATVQLLGEE424 pKa = 4.75PIAYY428 pKa = 9.85GKK430 pKa = 11.02VNGLDD435 pKa = 3.34SAAVLLVEE443 pKa = 4.35NTGGTGQFYY452 pKa = 10.31SLKK455 pKa = 9.95LVQRR459 pKa = 11.84IDD461 pKa = 3.54GEE463 pKa = 4.58AVNTASAFLGDD474 pKa = 3.91RR475 pKa = 11.84PRR477 pKa = 11.84IQNIVINEE485 pKa = 4.31DD486 pKa = 2.99GSITVDD492 pKa = 4.74LIQVGPNDD500 pKa = 4.33AFCCANTPMTVDD512 pKa = 4.15FVKK515 pKa = 10.41MGDD518 pKa = 3.28RR519 pKa = 11.84LIVQEE524 pKa = 4.95EE525 pKa = 3.96ISATIDD531 pKa = 3.42LAGLEE536 pKa = 4.67LPLIAAVKK544 pKa = 9.47PATAYY549 pKa = 9.47DD550 pKa = 3.52ASVPPGEE557 pKa = 4.31QGQPKK562 pKa = 9.12HH563 pKa = 6.38FSWVWTDD570 pKa = 4.65DD571 pKa = 3.38EE572 pKa = 4.59TAQGYY577 pKa = 9.74ISVYY581 pKa = 10.03PVEE584 pKa = 5.0AYY586 pKa = 9.62QAIWEE591 pKa = 4.25EE592 pKa = 3.87ADD594 pKa = 3.74DD595 pKa = 4.42PFVTEE600 pKa = 4.11TLAQLRR606 pKa = 11.84ALLEE610 pKa = 4.14EE611 pKa = 4.64QPANPPPPLPFLPPLPASNDD631 pKa = 2.8FAAQVRR637 pKa = 11.84YY638 pKa = 10.39LEE640 pKa = 4.49LADD643 pKa = 4.03GGVGVRR649 pKa = 11.84FIGRR653 pKa = 11.84FVQDD657 pKa = 2.81VAPIEE662 pKa = 4.32NYY664 pKa = 7.18QLRR667 pKa = 11.84YY668 pKa = 9.94LFQGLSADD676 pKa = 3.52GAYY679 pKa = 10.62LIVANLPVSTSALPAEE695 pKa = 4.38PQPLSGEE702 pKa = 4.3EE703 pKa = 3.78YY704 pKa = 9.29DD705 pKa = 3.98QFVAVYY711 pKa = 6.89TTYY714 pKa = 10.6LAEE717 pKa = 4.07MTTLFNGLTSTDD729 pKa = 3.96FTPDD733 pKa = 3.38LDD735 pKa = 4.25ALDD738 pKa = 5.93AILQSVTPKK747 pKa = 9.78RR748 pKa = 11.84TVNPLAPISLANMTINSEE766 pKa = 4.11LASDD770 pKa = 4.82GVAPLANGIYY780 pKa = 9.49TEE782 pKa = 4.58TVAPGSPGLIEE793 pKa = 4.62VRR795 pKa = 11.84LLPEE799 pKa = 3.91PMAYY803 pKa = 10.12GIINEE808 pKa = 4.24QEE810 pKa = 3.8VVAAIFTEE818 pKa = 4.12SDD820 pKa = 3.13GGSGIFVNLALIVAQDD836 pKa = 3.76GEE838 pKa = 4.57PVHH841 pKa = 6.32VASAPLGTRR850 pKa = 11.84VEE852 pKa = 4.2VQEE855 pKa = 5.6LSIADD860 pKa = 3.68NQIALTMLALGPDD873 pKa = 4.22DD874 pKa = 5.75PICCPSQRR882 pKa = 11.84VSHH885 pKa = 6.5AYY887 pKa = 9.5AWEE890 pKa = 4.52GDD892 pKa = 3.69VLALVEE898 pKa = 4.46EE899 pKa = 4.69RR900 pKa = 11.84VEE902 pKa = 4.28TTPEE906 pKa = 3.53ATLPLSNTSWAWVEE920 pKa = 4.28TVFNDD925 pKa = 3.75GTRR928 pKa = 11.84ITPATEE934 pKa = 4.09GAFTLTFTPEE944 pKa = 4.31GEE946 pKa = 4.34VSATTDD952 pKa = 3.45CNTFGGSYY960 pKa = 9.43RR961 pKa = 11.84AEE963 pKa = 4.18AGEE966 pKa = 3.9LAIEE970 pKa = 4.58FLTSTTIACPEE981 pKa = 4.09DD982 pKa = 3.51AQEE985 pKa = 4.16QEE987 pKa = 4.9FIADD991 pKa = 3.27VAAANGYY998 pKa = 6.87TVTEE1002 pKa = 4.33EE1003 pKa = 3.96GRR1005 pKa = 11.84LALLLPFNSGSVIFVPAAEE1024 pKa = 4.22VQVSDD1029 pKa = 3.96EE1030 pKa = 4.28AAAEE1034 pKa = 4.06ATTTDD1039 pKa = 3.35TAAEE1043 pKa = 4.34TTAEE1047 pKa = 4.32APLATTPVALPGTVWNWVRR1066 pKa = 11.84TQYY1069 pKa = 11.69SNGAIIAPSDD1079 pKa = 3.34PSLYY1083 pKa = 10.86VLTFGEE1089 pKa = 4.77DD1090 pKa = 3.18GTVNVQDD1097 pKa = 4.15DD1098 pKa = 4.57CNVVNGVFTAFADD1111 pKa = 3.68QALTLDD1117 pKa = 4.21LQTSTMAACAPDD1129 pKa = 3.38SLHH1132 pKa = 5.98YY1133 pKa = 10.56QFILDD1138 pKa = 3.94LSAAASYY1145 pKa = 9.01FFQDD1149 pKa = 2.42GRR1151 pKa = 11.84LFIDD1155 pKa = 4.15LKK1157 pKa = 10.93FDD1159 pKa = 3.24TGVIEE1164 pKa = 5.11FALADD1169 pKa = 3.53
MM1 pKa = 7.49LALALSALPTPLLAAPFQQEE21 pKa = 4.4VPVSDD26 pKa = 3.86GLTLEE31 pKa = 4.69ALANLTYY38 pKa = 10.37RR39 pKa = 11.84SEE41 pKa = 4.14LTPSGEE47 pKa = 3.74ITLVDD52 pKa = 3.94GRR54 pKa = 11.84YY55 pKa = 8.91EE56 pKa = 4.15DD57 pKa = 3.47PANRR61 pKa = 11.84VFAVLAAEE69 pKa = 4.25PMAFGTINGQDD80 pKa = 3.18AAAVLLAEE88 pKa = 4.33SSGAGSVFITLAIVVDD104 pKa = 3.99QDD106 pKa = 3.86GEE108 pKa = 4.42PVNVASTLLGDD119 pKa = 3.89RR120 pKa = 11.84VDD122 pKa = 4.17VYY124 pKa = 11.56ALEE127 pKa = 4.85IDD129 pKa = 4.19GSRR132 pKa = 11.84ILVDD136 pKa = 3.51MVRR139 pKa = 11.84QGPNDD144 pKa = 3.66PMCCPTEE151 pKa = 4.17VVRR154 pKa = 11.84QIYY157 pKa = 10.6ALGEE161 pKa = 4.03EE162 pKa = 4.55GLILAGVNLIGSGVQAALNVSPEE185 pKa = 3.72GGYY188 pKa = 7.15QTKK191 pKa = 10.2VMPATPYY198 pKa = 10.46AISALIPTGAPIHH211 pKa = 5.87TLLTFGDD218 pKa = 4.67DD219 pKa = 3.69DD220 pKa = 4.82PEE222 pKa = 4.56TVFLNGGPYY231 pKa = 8.91IAIYY235 pKa = 9.26PVQEE239 pKa = 4.46FVRR242 pKa = 11.84LWEE245 pKa = 4.01EE246 pKa = 4.25AGDD249 pKa = 3.72LTIANLVEE257 pKa = 3.93EE258 pKa = 5.15LKK260 pKa = 10.8RR261 pKa = 11.84ILTEE265 pKa = 4.09QPASLAAPLPIVPPADD281 pKa = 3.25VDD283 pKa = 4.17DD284 pKa = 5.35LVAQVVYY291 pKa = 11.05LEE293 pKa = 5.18GEE295 pKa = 4.24DD296 pKa = 3.63FTGVRR301 pKa = 11.84FIARR305 pKa = 11.84SVRR308 pKa = 11.84NGIVSADD315 pKa = 3.41GQLAYY320 pKa = 10.49HH321 pKa = 5.78FTGLTDD327 pKa = 3.72DD328 pKa = 3.5SRR330 pKa = 11.84YY331 pKa = 10.44LIAAQWPVEE340 pKa = 4.19TTAALTALDD349 pKa = 4.39SAPDD353 pKa = 4.02SAWTPALSSLDD364 pKa = 3.61VLIEE368 pKa = 3.97SLTIQGPEE376 pKa = 3.55WVTVEE381 pKa = 3.92EE382 pKa = 4.55LANLTYY388 pKa = 10.82DD389 pKa = 3.27SVMLEE394 pKa = 4.12KK395 pKa = 10.33PVTLSSGVYY404 pKa = 8.63TEE406 pKa = 4.29TVIPDD411 pKa = 3.53VPAGVATVQLLGEE424 pKa = 4.75PIAYY428 pKa = 9.85GKK430 pKa = 11.02VNGLDD435 pKa = 3.34SAAVLLVEE443 pKa = 4.35NTGGTGQFYY452 pKa = 10.31SLKK455 pKa = 9.95LVQRR459 pKa = 11.84IDD461 pKa = 3.54GEE463 pKa = 4.58AVNTASAFLGDD474 pKa = 3.91RR475 pKa = 11.84PRR477 pKa = 11.84IQNIVINEE485 pKa = 4.31DD486 pKa = 2.99GSITVDD492 pKa = 4.74LIQVGPNDD500 pKa = 4.33AFCCANTPMTVDD512 pKa = 4.15FVKK515 pKa = 10.41MGDD518 pKa = 3.28RR519 pKa = 11.84LIVQEE524 pKa = 4.95EE525 pKa = 3.96ISATIDD531 pKa = 3.42LAGLEE536 pKa = 4.67LPLIAAVKK544 pKa = 9.47PATAYY549 pKa = 9.47DD550 pKa = 3.52ASVPPGEE557 pKa = 4.31QGQPKK562 pKa = 9.12HH563 pKa = 6.38FSWVWTDD570 pKa = 4.65DD571 pKa = 3.38EE572 pKa = 4.59TAQGYY577 pKa = 9.74ISVYY581 pKa = 10.03PVEE584 pKa = 5.0AYY586 pKa = 9.62QAIWEE591 pKa = 4.25EE592 pKa = 3.87ADD594 pKa = 3.74DD595 pKa = 4.42PFVTEE600 pKa = 4.11TLAQLRR606 pKa = 11.84ALLEE610 pKa = 4.14EE611 pKa = 4.64QPANPPPPLPFLPPLPASNDD631 pKa = 2.8FAAQVRR637 pKa = 11.84YY638 pKa = 10.39LEE640 pKa = 4.49LADD643 pKa = 4.03GGVGVRR649 pKa = 11.84FIGRR653 pKa = 11.84FVQDD657 pKa = 2.81VAPIEE662 pKa = 4.32NYY664 pKa = 7.18QLRR667 pKa = 11.84YY668 pKa = 9.94LFQGLSADD676 pKa = 3.52GAYY679 pKa = 10.62LIVANLPVSTSALPAEE695 pKa = 4.38PQPLSGEE702 pKa = 4.3EE703 pKa = 3.78YY704 pKa = 9.29DD705 pKa = 3.98QFVAVYY711 pKa = 6.89TTYY714 pKa = 10.6LAEE717 pKa = 4.07MTTLFNGLTSTDD729 pKa = 3.96FTPDD733 pKa = 3.38LDD735 pKa = 4.25ALDD738 pKa = 5.93AILQSVTPKK747 pKa = 9.78RR748 pKa = 11.84TVNPLAPISLANMTINSEE766 pKa = 4.11LASDD770 pKa = 4.82GVAPLANGIYY780 pKa = 9.49TEE782 pKa = 4.58TVAPGSPGLIEE793 pKa = 4.62VRR795 pKa = 11.84LLPEE799 pKa = 3.91PMAYY803 pKa = 10.12GIINEE808 pKa = 4.24QEE810 pKa = 3.8VVAAIFTEE818 pKa = 4.12SDD820 pKa = 3.13GGSGIFVNLALIVAQDD836 pKa = 3.76GEE838 pKa = 4.57PVHH841 pKa = 6.32VASAPLGTRR850 pKa = 11.84VEE852 pKa = 4.2VQEE855 pKa = 5.6LSIADD860 pKa = 3.68NQIALTMLALGPDD873 pKa = 4.22DD874 pKa = 5.75PICCPSQRR882 pKa = 11.84VSHH885 pKa = 6.5AYY887 pKa = 9.5AWEE890 pKa = 4.52GDD892 pKa = 3.69VLALVEE898 pKa = 4.46EE899 pKa = 4.69RR900 pKa = 11.84VEE902 pKa = 4.28TTPEE906 pKa = 3.53ATLPLSNTSWAWVEE920 pKa = 4.28TVFNDD925 pKa = 3.75GTRR928 pKa = 11.84ITPATEE934 pKa = 4.09GAFTLTFTPEE944 pKa = 4.31GEE946 pKa = 4.34VSATTDD952 pKa = 3.45CNTFGGSYY960 pKa = 9.43RR961 pKa = 11.84AEE963 pKa = 4.18AGEE966 pKa = 3.9LAIEE970 pKa = 4.58FLTSTTIACPEE981 pKa = 4.09DD982 pKa = 3.51AQEE985 pKa = 4.16QEE987 pKa = 4.9FIADD991 pKa = 3.27VAAANGYY998 pKa = 6.87TVTEE1002 pKa = 4.33EE1003 pKa = 3.96GRR1005 pKa = 11.84LALLLPFNSGSVIFVPAAEE1024 pKa = 4.22VQVSDD1029 pKa = 3.96EE1030 pKa = 4.28AAAEE1034 pKa = 4.06ATTTDD1039 pKa = 3.35TAAEE1043 pKa = 4.34TTAEE1047 pKa = 4.32APLATTPVALPGTVWNWVRR1066 pKa = 11.84TQYY1069 pKa = 11.69SNGAIIAPSDD1079 pKa = 3.34PSLYY1083 pKa = 10.86VLTFGEE1089 pKa = 4.77DD1090 pKa = 3.18GTVNVQDD1097 pKa = 4.15DD1098 pKa = 4.57CNVVNGVFTAFADD1111 pKa = 3.68QALTLDD1117 pKa = 4.21LQTSTMAACAPDD1129 pKa = 3.38SLHH1132 pKa = 5.98YY1133 pKa = 10.56QFILDD1138 pKa = 3.94LSAAASYY1145 pKa = 9.01FFQDD1149 pKa = 2.42GRR1151 pKa = 11.84LFIDD1155 pKa = 4.15LKK1157 pKa = 10.93FDD1159 pKa = 3.24TGVIEE1164 pKa = 5.11FALADD1169 pKa = 3.53
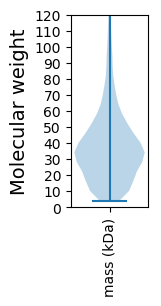
Molecular weight: 124.09 kDa
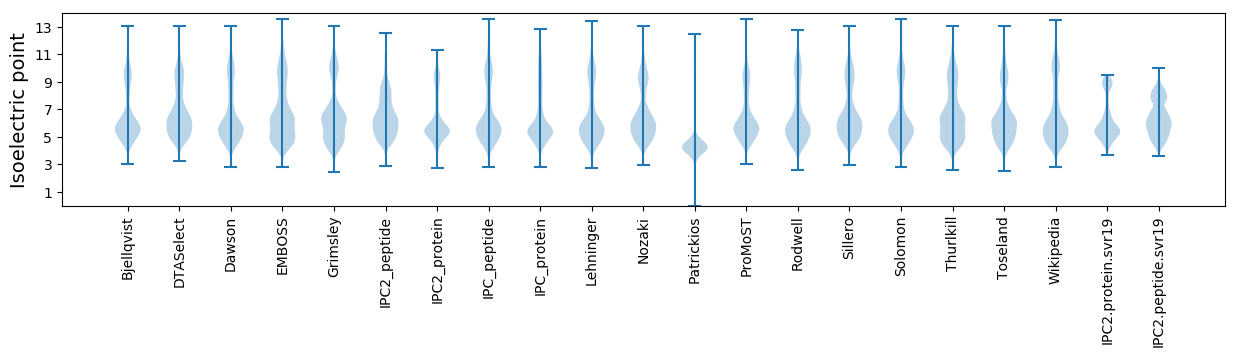
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0HZV6|I0HZV6_CALAS ATP synthase epsilon chain OS=Caldilinea aerophila (strain DSM 14535 / JCM 11387 / NBRC 104270 / STL-6-O1) OX=926550 GN=atpC PE=3 SV=1
MM1 pKa = 6.75ATKK4 pKa = 9.53RR5 pKa = 11.84TWQPKK10 pKa = 6.57VRR12 pKa = 11.84KK13 pKa = 9.2RR14 pKa = 11.84LKK16 pKa = 8.26THH18 pKa = 5.87GFRR21 pKa = 11.84IRR23 pKa = 11.84MSTPGGRR30 pKa = 11.84KK31 pKa = 7.15VLKK34 pKa = 10.05RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84LKK39 pKa = 10.44GRR41 pKa = 11.84ARR43 pKa = 11.84LTVQLSHH50 pKa = 7.07RR51 pKa = 11.84KK52 pKa = 9.64AII54 pKa = 3.9
MM1 pKa = 6.75ATKK4 pKa = 9.53RR5 pKa = 11.84TWQPKK10 pKa = 6.57VRR12 pKa = 11.84KK13 pKa = 9.2RR14 pKa = 11.84LKK16 pKa = 8.26THH18 pKa = 5.87GFRR21 pKa = 11.84IRR23 pKa = 11.84MSTPGGRR30 pKa = 11.84KK31 pKa = 7.15VLKK34 pKa = 10.05RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84LKK39 pKa = 10.44GRR41 pKa = 11.84ARR43 pKa = 11.84LTVQLSHH50 pKa = 7.07RR51 pKa = 11.84KK52 pKa = 9.64AII54 pKa = 3.9
Molecular weight: 6.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1491047 |
37 |
5788 |
363.9 |
40.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.222 ± 0.05 | 0.832 ± 0.013 |
4.876 ± 0.028 | 6.206 ± 0.04 |
3.761 ± 0.029 | 7.725 ± 0.035 |
2.127 ± 0.021 | 5.351 ± 0.029 |
2.399 ± 0.027 | 10.988 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.245 ± 0.018 | 2.889 ± 0.024 |
5.755 ± 0.036 | 3.856 ± 0.024 |
7.023 ± 0.039 | 4.936 ± 0.025 |
5.498 ± 0.048 | 7.66 ± 0.031 |
1.76 ± 0.021 | 2.891 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
