
Carrot mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
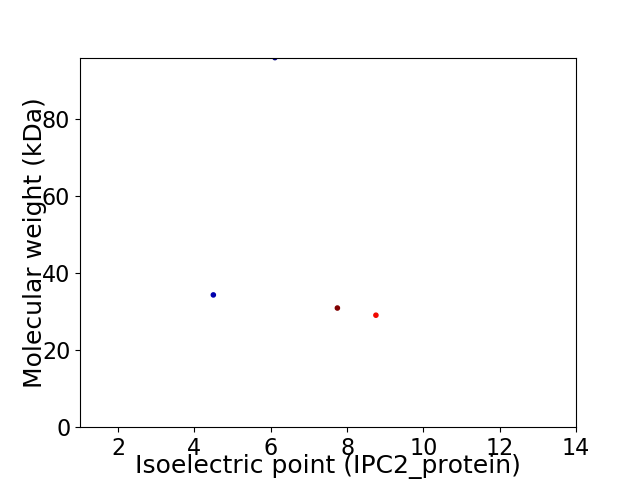
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6UQC5|B6UQC5_9TOMB RNA-directed RNA polymerase OS=Carrot mottle virus OX=68033 PE=4 SV=2
MM1 pKa = 7.43SFLKK5 pKa = 10.63RR6 pKa = 11.84ILKK9 pKa = 10.14HH10 pKa = 6.43LDD12 pKa = 3.45SQFSPAPGVYY22 pKa = 10.11NRR24 pKa = 11.84DD25 pKa = 3.01VCAEE29 pKa = 4.19LYY31 pKa = 9.13GAEE34 pKa = 3.97WGVLITQEE42 pKa = 3.8RR43 pKa = 11.84MTRR46 pKa = 11.84ALSVEE51 pKa = 3.85HH52 pKa = 6.09EE53 pKa = 3.75RR54 pKa = 11.84WYY56 pKa = 9.42TGRR59 pKa = 11.84VEE61 pKa = 4.17TDD63 pKa = 2.98VYY65 pKa = 11.0IPVAQYY71 pKa = 10.75VDD73 pKa = 3.6PRR75 pKa = 11.84DD76 pKa = 3.8VEE78 pKa = 4.58AEE80 pKa = 3.98PTSQQGNSTGVDD92 pKa = 3.16NPVNNCSGTVPTMPATEE109 pKa = 4.51VIPVDD114 pKa = 3.45GLGGRR119 pKa = 11.84RR120 pKa = 11.84VGVVPAPEE128 pKa = 4.37GPIPCEE134 pKa = 3.83GNVYY138 pKa = 10.52AILRR142 pKa = 11.84MADD145 pKa = 3.35PTADD149 pKa = 3.42ASEE152 pKa = 4.56SSDD155 pKa = 4.65DD156 pKa = 5.59EE157 pKa = 5.23EE158 pKa = 4.78DD159 pKa = 3.79TCCPAYY165 pKa = 9.79IHH167 pKa = 6.62AASNSVAEE175 pKa = 4.53HH176 pKa = 6.24SANAVPPPMEE186 pKa = 4.38GVHH189 pKa = 6.69PGTSPDD195 pKa = 3.64GVPSSSSAVGISLAVDD211 pKa = 3.44VTHH214 pKa = 6.72VNTVDD219 pKa = 4.04ASTEE223 pKa = 3.94MPTKK227 pKa = 10.76VEE229 pKa = 4.29GPDD232 pKa = 3.42EE233 pKa = 4.5GVSTSPLQEE242 pKa = 4.99AGPSMAFVDD251 pKa = 4.29ASSVAMEE258 pKa = 3.81LRR260 pKa = 11.84ARR262 pKa = 11.84FGYY265 pKa = 10.01RR266 pKa = 11.84SKK268 pKa = 11.41NPANLEE274 pKa = 3.82LGGRR278 pKa = 11.84VARR281 pKa = 11.84DD282 pKa = 3.21ILKK285 pKa = 9.97GCGAVRR291 pKa = 11.84SEE293 pKa = 4.02NYY295 pKa = 10.27YY296 pKa = 10.39LAQLAVQLWFAPTLLDD312 pKa = 3.52LVLRR316 pKa = 11.84GPVQDD321 pKa = 4.13FCC323 pKa = 6.41
MM1 pKa = 7.43SFLKK5 pKa = 10.63RR6 pKa = 11.84ILKK9 pKa = 10.14HH10 pKa = 6.43LDD12 pKa = 3.45SQFSPAPGVYY22 pKa = 10.11NRR24 pKa = 11.84DD25 pKa = 3.01VCAEE29 pKa = 4.19LYY31 pKa = 9.13GAEE34 pKa = 3.97WGVLITQEE42 pKa = 3.8RR43 pKa = 11.84MTRR46 pKa = 11.84ALSVEE51 pKa = 3.85HH52 pKa = 6.09EE53 pKa = 3.75RR54 pKa = 11.84WYY56 pKa = 9.42TGRR59 pKa = 11.84VEE61 pKa = 4.17TDD63 pKa = 2.98VYY65 pKa = 11.0IPVAQYY71 pKa = 10.75VDD73 pKa = 3.6PRR75 pKa = 11.84DD76 pKa = 3.8VEE78 pKa = 4.58AEE80 pKa = 3.98PTSQQGNSTGVDD92 pKa = 3.16NPVNNCSGTVPTMPATEE109 pKa = 4.51VIPVDD114 pKa = 3.45GLGGRR119 pKa = 11.84RR120 pKa = 11.84VGVVPAPEE128 pKa = 4.37GPIPCEE134 pKa = 3.83GNVYY138 pKa = 10.52AILRR142 pKa = 11.84MADD145 pKa = 3.35PTADD149 pKa = 3.42ASEE152 pKa = 4.56SSDD155 pKa = 4.65DD156 pKa = 5.59EE157 pKa = 5.23EE158 pKa = 4.78DD159 pKa = 3.79TCCPAYY165 pKa = 9.79IHH167 pKa = 6.62AASNSVAEE175 pKa = 4.53HH176 pKa = 6.24SANAVPPPMEE186 pKa = 4.38GVHH189 pKa = 6.69PGTSPDD195 pKa = 3.64GVPSSSSAVGISLAVDD211 pKa = 3.44VTHH214 pKa = 6.72VNTVDD219 pKa = 4.04ASTEE223 pKa = 3.94MPTKK227 pKa = 10.76VEE229 pKa = 4.29GPDD232 pKa = 3.42EE233 pKa = 4.5GVSTSPLQEE242 pKa = 4.99AGPSMAFVDD251 pKa = 4.29ASSVAMEE258 pKa = 3.81LRR260 pKa = 11.84ARR262 pKa = 11.84FGYY265 pKa = 10.01RR266 pKa = 11.84SKK268 pKa = 11.41NPANLEE274 pKa = 3.82LGGRR278 pKa = 11.84VARR281 pKa = 11.84DD282 pKa = 3.21ILKK285 pKa = 9.97GCGAVRR291 pKa = 11.84SEE293 pKa = 4.02NYY295 pKa = 10.27YY296 pKa = 10.39LAQLAVQLWFAPTLLDD312 pKa = 3.52LVLRR316 pKa = 11.84GPVQDD321 pKa = 4.13FCC323 pKa = 6.41
Molecular weight: 34.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6UQC7|B6UQC7_9TOMB Cell-to-cell movement protein OS=Carrot mottle virus OX=68033 PE=4 SV=1
MM1 pKa = 7.43TFRR4 pKa = 11.84IIYY7 pKa = 9.99DD8 pKa = 4.09GIEE11 pKa = 3.54QDD13 pKa = 3.35SIYY16 pKa = 10.84NRR18 pKa = 11.84RR19 pKa = 11.84THH21 pKa = 5.95QPPARR26 pKa = 11.84RR27 pKa = 11.84NNSGRR32 pKa = 11.84TARR35 pKa = 11.84ARR37 pKa = 11.84AWDD40 pKa = 3.87YY41 pKa = 11.65GPSPWHH47 pKa = 5.91QYY49 pKa = 10.72AYY51 pKa = 9.27PHH53 pKa = 6.54SALPPPAPAYY63 pKa = 10.02FEE65 pKa = 5.01PLFQRR70 pKa = 11.84PLAYY74 pKa = 10.29QEE76 pKa = 4.47DD77 pKa = 4.42RR78 pKa = 11.84GHH80 pKa = 6.03AVYY83 pKa = 10.57RR84 pKa = 11.84EE85 pKa = 4.03ARR87 pKa = 11.84HH88 pKa = 4.98SVRR91 pKa = 11.84SARR94 pKa = 11.84ARR96 pKa = 11.84HH97 pKa = 4.83VAGTSSDD104 pKa = 2.74LGARR108 pKa = 11.84HH109 pKa = 6.08RR110 pKa = 11.84PPRR113 pKa = 11.84SGTDD117 pKa = 2.96RR118 pKa = 11.84DD119 pKa = 4.15GPEE122 pKa = 3.75GVYY125 pKa = 9.9TLVLGTPAGRR135 pKa = 11.84FLSEE139 pKa = 4.36LFHH142 pKa = 7.85SFEE145 pKa = 4.49RR146 pKa = 11.84LSHH149 pKa = 6.18EE150 pKa = 4.61CPSVLQSSNPVRR162 pKa = 11.84GGTTGARR169 pKa = 11.84GEE171 pKa = 4.18RR172 pKa = 11.84LLSLLHH178 pKa = 5.86VAASHH183 pKa = 6.19RR184 pKa = 11.84GEE186 pKa = 4.44GAVLPADD193 pKa = 3.82TTGGCGDD200 pKa = 4.04TTSQHH205 pKa = 6.88PIPTPGLEE213 pKa = 4.11PAHH216 pKa = 6.28AVPTRR221 pKa = 11.84RR222 pKa = 11.84DD223 pKa = 3.22DD224 pKa = 3.63EE225 pKa = 5.17RR226 pKa = 11.84YY227 pKa = 10.07GNHH230 pKa = 6.97DD231 pKa = 3.45QGQSIHH237 pKa = 6.87ALPAPHH243 pKa = 7.14LCGKK247 pKa = 8.63VHH249 pKa = 6.84AVGGPCPTRR258 pKa = 11.84EE259 pKa = 4.13SEE261 pKa = 4.13VKK263 pKa = 10.52YY264 pKa = 10.64RR265 pKa = 3.79
MM1 pKa = 7.43TFRR4 pKa = 11.84IIYY7 pKa = 9.99DD8 pKa = 4.09GIEE11 pKa = 3.54QDD13 pKa = 3.35SIYY16 pKa = 10.84NRR18 pKa = 11.84RR19 pKa = 11.84THH21 pKa = 5.95QPPARR26 pKa = 11.84RR27 pKa = 11.84NNSGRR32 pKa = 11.84TARR35 pKa = 11.84ARR37 pKa = 11.84AWDD40 pKa = 3.87YY41 pKa = 11.65GPSPWHH47 pKa = 5.91QYY49 pKa = 10.72AYY51 pKa = 9.27PHH53 pKa = 6.54SALPPPAPAYY63 pKa = 10.02FEE65 pKa = 5.01PLFQRR70 pKa = 11.84PLAYY74 pKa = 10.29QEE76 pKa = 4.47DD77 pKa = 4.42RR78 pKa = 11.84GHH80 pKa = 6.03AVYY83 pKa = 10.57RR84 pKa = 11.84EE85 pKa = 4.03ARR87 pKa = 11.84HH88 pKa = 4.98SVRR91 pKa = 11.84SARR94 pKa = 11.84ARR96 pKa = 11.84HH97 pKa = 4.83VAGTSSDD104 pKa = 2.74LGARR108 pKa = 11.84HH109 pKa = 6.08RR110 pKa = 11.84PPRR113 pKa = 11.84SGTDD117 pKa = 2.96RR118 pKa = 11.84DD119 pKa = 4.15GPEE122 pKa = 3.75GVYY125 pKa = 9.9TLVLGTPAGRR135 pKa = 11.84FLSEE139 pKa = 4.36LFHH142 pKa = 7.85SFEE145 pKa = 4.49RR146 pKa = 11.84LSHH149 pKa = 6.18EE150 pKa = 4.61CPSVLQSSNPVRR162 pKa = 11.84GGTTGARR169 pKa = 11.84GEE171 pKa = 4.18RR172 pKa = 11.84LLSLLHH178 pKa = 5.86VAASHH183 pKa = 6.19RR184 pKa = 11.84GEE186 pKa = 4.44GAVLPADD193 pKa = 3.82TTGGCGDD200 pKa = 4.04TTSQHH205 pKa = 6.88PIPTPGLEE213 pKa = 4.11PAHH216 pKa = 6.28AVPTRR221 pKa = 11.84RR222 pKa = 11.84DD223 pKa = 3.22DD224 pKa = 3.63EE225 pKa = 5.17RR226 pKa = 11.84YY227 pKa = 10.07GNHH230 pKa = 6.97DD231 pKa = 3.45QGQSIHH237 pKa = 6.87ALPAPHH243 pKa = 7.14LCGKK247 pKa = 8.63VHH249 pKa = 6.84AVGGPCPTRR258 pKa = 11.84EE259 pKa = 4.13SEE261 pKa = 4.13VKK263 pKa = 10.52YY264 pKa = 10.64RR265 pKa = 3.79
Molecular weight: 29.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1737 |
265 |
870 |
434.3 |
47.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.002 ± 0.812 | 1.957 ± 0.327 |
5.584 ± 0.558 | 5.527 ± 0.455 |
3.224 ± 0.654 | 7.83 ± 0.53 |
2.879 ± 0.921 | 3.8 ± 0.591 |
3.224 ± 1.197 | 7.599 ± 0.697 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.469 | 3.109 ± 0.307 |
7.945 ± 0.75 | 3.627 ± 0.307 |
6.908 ± 1.099 | 7.772 ± 0.298 |
5.815 ± 0.811 | 8.52 ± 1.091 |
1.094 ± 0.13 | 3.282 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
