
Ascidiaceihabitans donghaensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Ascidiaceihabitans
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
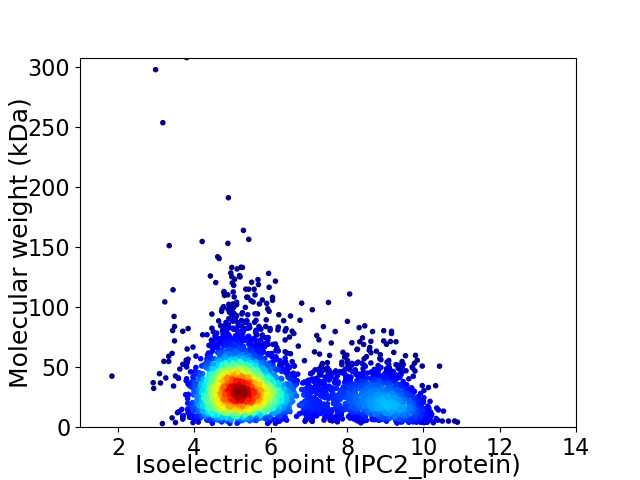
Virtual 2D-PAGE plot for 4027 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R8BFQ3|A0A2R8BFQ3_9RHOB Uncharacterized protein OS=Ascidiaceihabitans donghaensis OX=1510460 GN=ASD8599_02589 PE=4 SV=1
MM1 pKa = 7.48GKK3 pKa = 9.7FLAVLLVGSAFGAGIFMYY21 pKa = 10.1YY22 pKa = 9.36LQVYY26 pKa = 8.3GFYY29 pKa = 10.92DD30 pKa = 3.79EE31 pKa = 5.55VVGGDD36 pKa = 3.51VQLVSVVDD44 pKa = 4.57DD45 pKa = 4.06LPEE48 pKa = 4.52TISFDD53 pKa = 3.25NLQAIDD59 pKa = 4.4ADD61 pKa = 4.25SSPLRR66 pKa = 11.84YY67 pKa = 9.08RR68 pKa = 11.84ACFTTDD74 pKa = 3.18LSLSLLSEE82 pKa = 4.49TYY84 pKa = 10.72VGLEE88 pKa = 3.74TATPRR93 pKa = 11.84NAPEE97 pKa = 3.81WFDD100 pKa = 4.19CFDD103 pKa = 3.82AATIAAEE110 pKa = 4.32LEE112 pKa = 4.23AGTAIPFLGGKK123 pKa = 8.5NVEE126 pKa = 4.41FGVDD130 pKa = 3.57RR131 pKa = 11.84IVAITEE137 pKa = 3.95DD138 pKa = 3.03GRR140 pKa = 11.84GYY142 pKa = 10.46VWHH145 pKa = 7.71DD146 pKa = 4.17LNDD149 pKa = 4.55CGDD152 pKa = 3.41KK153 pKa = 10.87AYY155 pKa = 10.72DD156 pKa = 3.5GTIVGEE162 pKa = 4.01EE163 pKa = 4.14CPRR166 pKa = 11.84RR167 pKa = 11.84EE168 pKa = 4.07TEE170 pKa = 3.69
MM1 pKa = 7.48GKK3 pKa = 9.7FLAVLLVGSAFGAGIFMYY21 pKa = 10.1YY22 pKa = 9.36LQVYY26 pKa = 8.3GFYY29 pKa = 10.92DD30 pKa = 3.79EE31 pKa = 5.55VVGGDD36 pKa = 3.51VQLVSVVDD44 pKa = 4.57DD45 pKa = 4.06LPEE48 pKa = 4.52TISFDD53 pKa = 3.25NLQAIDD59 pKa = 4.4ADD61 pKa = 4.25SSPLRR66 pKa = 11.84YY67 pKa = 9.08RR68 pKa = 11.84ACFTTDD74 pKa = 3.18LSLSLLSEE82 pKa = 4.49TYY84 pKa = 10.72VGLEE88 pKa = 3.74TATPRR93 pKa = 11.84NAPEE97 pKa = 3.81WFDD100 pKa = 4.19CFDD103 pKa = 3.82AATIAAEE110 pKa = 4.32LEE112 pKa = 4.23AGTAIPFLGGKK123 pKa = 8.5NVEE126 pKa = 4.41FGVDD130 pKa = 3.57RR131 pKa = 11.84IVAITEE137 pKa = 3.95DD138 pKa = 3.03GRR140 pKa = 11.84GYY142 pKa = 10.46VWHH145 pKa = 7.71DD146 pKa = 4.17LNDD149 pKa = 4.55CGDD152 pKa = 3.41KK153 pKa = 10.87AYY155 pKa = 10.72DD156 pKa = 3.5GTIVGEE162 pKa = 4.01EE163 pKa = 4.14CPRR166 pKa = 11.84RR167 pKa = 11.84EE168 pKa = 4.07TEE170 pKa = 3.69
Molecular weight: 18.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R8BFM7|A0A2R8BFM7_9RHOB NHase_beta domain-containing protein OS=Ascidiaceihabitans donghaensis OX=1510460 GN=ASD8599_02665 PE=4 SV=1
MM1 pKa = 7.68PLLRR5 pKa = 11.84INATSEE11 pKa = 4.17GPQLHH16 pKa = 6.83GSPVPARR23 pKa = 11.84RR24 pKa = 11.84VIQTAARR31 pKa = 11.84SKK33 pKa = 10.29GPVIIMVHH41 pKa = 5.19GFKK44 pKa = 10.49YY45 pKa = 10.68DD46 pKa = 3.38PEE48 pKa = 4.44TPRR51 pKa = 11.84FCPHH55 pKa = 6.53ISIFDD60 pKa = 3.7APSGRR65 pKa = 11.84KK66 pKa = 8.84RR67 pKa = 11.84DD68 pKa = 3.42SWPAALGFAGRR79 pKa = 11.84DD80 pKa = 3.75PNEE83 pKa = 3.91GLAIAFAWRR92 pKa = 11.84ARR94 pKa = 11.84GTLWHH99 pKa = 7.12ALRR102 pKa = 11.84TAAQAGQNLAQVIAIVKK119 pKa = 7.94RR120 pKa = 11.84TAPARR125 pKa = 11.84AVHH128 pKa = 6.15VICHH132 pKa = 5.32SMGSEE137 pKa = 3.99VLFEE141 pKa = 4.71ALHH144 pKa = 6.15HH145 pKa = 6.41LPAHH149 pKa = 6.55SIQRR153 pKa = 11.84AIILTAASYY162 pKa = 10.28QSRR165 pKa = 11.84ASAALHH171 pKa = 5.97TDD173 pKa = 3.24AGQTLEE179 pKa = 4.61LFNITSRR186 pKa = 11.84EE187 pKa = 3.63NDD189 pKa = 3.27LFDD192 pKa = 5.55AGFEE196 pKa = 4.23TLIKK200 pKa = 10.5AATPSDD206 pKa = 3.57RR207 pKa = 11.84AVGAGLCAPNAVTIEE222 pKa = 3.99IDD224 pKa = 4.3CDD226 pKa = 3.41LTLQRR231 pKa = 11.84LRR233 pKa = 11.84LTGADD238 pKa = 3.03IAPAARR244 pKa = 11.84RR245 pKa = 11.84ISHH248 pKa = 6.39WSAYY252 pKa = 7.31MRR254 pKa = 11.84PGLMPFYY261 pKa = 10.65NSLLRR266 pKa = 11.84RR267 pKa = 11.84PSEE270 pKa = 4.14TPLAALRR277 pKa = 11.84HH278 pKa = 5.54ALPLVGSQRR287 pKa = 11.84FARR290 pKa = 11.84LLPFMHH296 pKa = 6.95KK297 pKa = 9.95PQAGLPAPLL306 pKa = 4.38
MM1 pKa = 7.68PLLRR5 pKa = 11.84INATSEE11 pKa = 4.17GPQLHH16 pKa = 6.83GSPVPARR23 pKa = 11.84RR24 pKa = 11.84VIQTAARR31 pKa = 11.84SKK33 pKa = 10.29GPVIIMVHH41 pKa = 5.19GFKK44 pKa = 10.49YY45 pKa = 10.68DD46 pKa = 3.38PEE48 pKa = 4.44TPRR51 pKa = 11.84FCPHH55 pKa = 6.53ISIFDD60 pKa = 3.7APSGRR65 pKa = 11.84KK66 pKa = 8.84RR67 pKa = 11.84DD68 pKa = 3.42SWPAALGFAGRR79 pKa = 11.84DD80 pKa = 3.75PNEE83 pKa = 3.91GLAIAFAWRR92 pKa = 11.84ARR94 pKa = 11.84GTLWHH99 pKa = 7.12ALRR102 pKa = 11.84TAAQAGQNLAQVIAIVKK119 pKa = 7.94RR120 pKa = 11.84TAPARR125 pKa = 11.84AVHH128 pKa = 6.15VICHH132 pKa = 5.32SMGSEE137 pKa = 3.99VLFEE141 pKa = 4.71ALHH144 pKa = 6.15HH145 pKa = 6.41LPAHH149 pKa = 6.55SIQRR153 pKa = 11.84AIILTAASYY162 pKa = 10.28QSRR165 pKa = 11.84ASAALHH171 pKa = 5.97TDD173 pKa = 3.24AGQTLEE179 pKa = 4.61LFNITSRR186 pKa = 11.84EE187 pKa = 3.63NDD189 pKa = 3.27LFDD192 pKa = 5.55AGFEE196 pKa = 4.23TLIKK200 pKa = 10.5AATPSDD206 pKa = 3.57RR207 pKa = 11.84AVGAGLCAPNAVTIEE222 pKa = 3.99IDD224 pKa = 4.3CDD226 pKa = 3.41LTLQRR231 pKa = 11.84LRR233 pKa = 11.84LTGADD238 pKa = 3.03IAPAARR244 pKa = 11.84RR245 pKa = 11.84ISHH248 pKa = 6.39WSAYY252 pKa = 7.31MRR254 pKa = 11.84PGLMPFYY261 pKa = 10.65NSLLRR266 pKa = 11.84RR267 pKa = 11.84PSEE270 pKa = 4.14TPLAALRR277 pKa = 11.84HH278 pKa = 5.54ALPLVGSQRR287 pKa = 11.84FARR290 pKa = 11.84LLPFMHH296 pKa = 6.95KK297 pKa = 9.95PQAGLPAPLL306 pKa = 4.38
Molecular weight: 33.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1246692 |
29 |
3150 |
309.6 |
33.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.687 ± 0.048 | 0.94 ± 0.012 |
6.522 ± 0.038 | 5.223 ± 0.039 |
3.921 ± 0.024 | 8.532 ± 0.043 |
2.175 ± 0.022 | 5.258 ± 0.026 |
3.682 ± 0.028 | 9.531 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.006 ± 0.022 | 3.034 ± 0.03 |
4.791 ± 0.025 | 3.527 ± 0.022 |
5.807 ± 0.039 | 5.376 ± 0.028 |
5.934 ± 0.041 | 7.383 ± 0.027 |
1.391 ± 0.018 | 2.281 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
