
Beihai sea slater virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
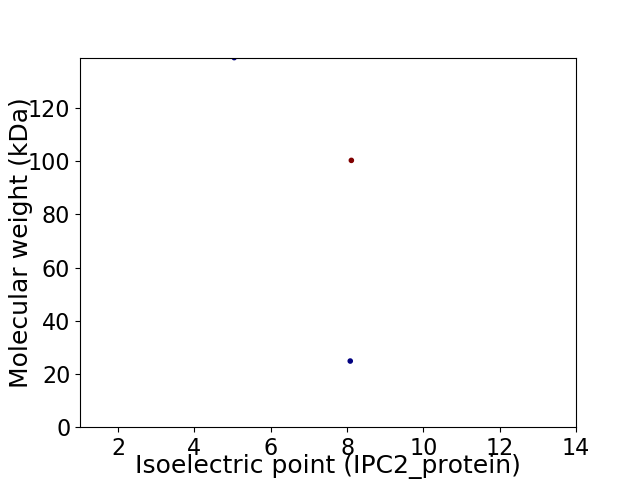
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
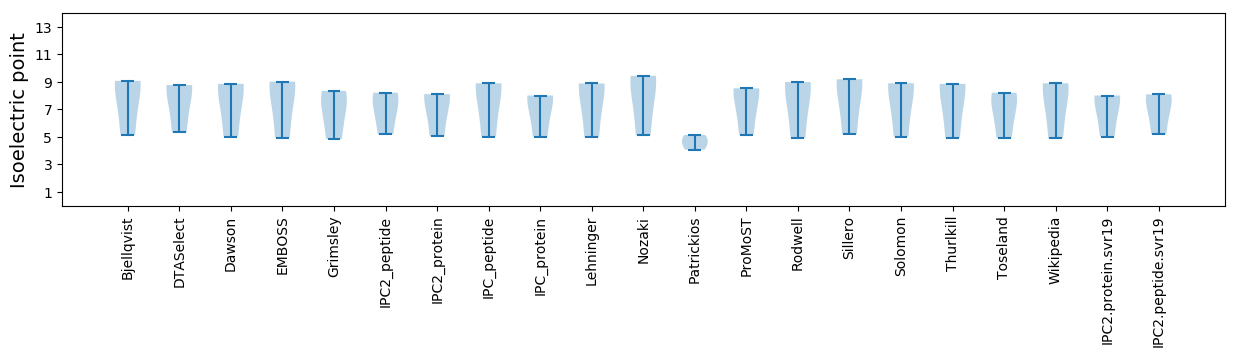
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF04|A0A1L3KF04_9VIRU RNA-directed RNA polymerase OS=Beihai sea slater virus 3 OX=1922659 PE=4 SV=1
MM1 pKa = 7.62AFLIPALVGGIEE13 pKa = 3.92AAVAGGGAVLGNKK26 pKa = 8.94LAHH29 pKa = 6.36KK30 pKa = 10.53VVGDD34 pKa = 4.18APADD38 pKa = 3.44LEE40 pKa = 4.76GIMEE44 pKa = 4.26VMQFLDD50 pKa = 3.8PSQNPSSMPRR60 pKa = 11.84MLQSQEE66 pKa = 4.28DD67 pKa = 3.94LSSALEE73 pKa = 3.94TRR75 pKa = 11.84LNIQGLDD82 pKa = 3.54NEE84 pKa = 4.76KK85 pKa = 9.24KK86 pKa = 8.89TKK88 pKa = 9.89FVSNLQALKK97 pKa = 10.57GQADD101 pKa = 4.16LALKK105 pKa = 10.56DD106 pKa = 3.82LLRR109 pKa = 11.84LKK111 pKa = 9.46STQGLSDD118 pKa = 3.29ATAVMNQAPNLIQNQPSLTSNPVPQAQVVTSPGLNVQARR157 pKa = 11.84EE158 pKa = 4.25APPTLQTLIQNLQAQAVPQSQVQDD182 pKa = 3.48SSALTRR188 pKa = 11.84NEE190 pKa = 3.66PSTTFSAVYY199 pKa = 9.34KK200 pKa = 9.87YY201 pKa = 10.1PVSRR205 pKa = 11.84LPLQASQPISKK216 pKa = 10.22NVLNQGTVIPRR227 pKa = 11.84PQVEE231 pKa = 4.69GTSSSQGTTNLRR243 pKa = 11.84TTASDD248 pKa = 3.52TVQGVGQALKK258 pKa = 10.68GAASDD263 pKa = 3.57AGKK266 pKa = 10.61VAVTALGNAAAEE278 pKa = 4.29VGKK281 pKa = 10.77GVLSHH286 pKa = 6.96LGKK289 pKa = 10.41LAAAKK294 pKa = 10.24IGDD297 pKa = 3.99FSSILAKK304 pKa = 10.48KK305 pKa = 9.95IVGDD309 pKa = 3.8AVPANDD315 pKa = 3.92HH316 pKa = 7.47RR317 pKa = 11.84IGTWMRR323 pKa = 11.84SEE325 pKa = 4.24KK326 pKa = 10.7VVGDD330 pKa = 4.06AEE332 pKa = 4.79VSTKK336 pKa = 10.94DD337 pKa = 3.55NPKK340 pKa = 10.7LMALLQQMANGGSAPKK356 pKa = 10.37SGEE359 pKa = 3.95DD360 pKa = 3.27KK361 pKa = 10.88TDD363 pKa = 3.15GKK365 pKa = 10.7IVGDD369 pKa = 3.91APPGVMEE376 pKa = 4.71KK377 pKa = 10.17QDD379 pKa = 3.4EE380 pKa = 4.68TMTDD384 pKa = 3.39EE385 pKa = 4.35QVAAAAGNLQSSPSLALMSHH405 pKa = 6.31IQLDD409 pKa = 4.04EE410 pKa = 4.03QMLEE414 pKa = 3.91SGKK417 pKa = 10.24VVEE420 pKa = 4.86FTSDD424 pKa = 2.87VSEE427 pKa = 4.33AARR430 pKa = 11.84MIARR434 pKa = 11.84LGEE437 pKa = 4.12EE438 pKa = 3.88VSGVFKK444 pKa = 9.84TEE446 pKa = 3.66RR447 pKa = 11.84LISTRR452 pKa = 11.84TQVGNQYY459 pKa = 10.05FWNTYY464 pKa = 6.58FTLRR468 pKa = 11.84NPRR471 pKa = 11.84LDD473 pKa = 3.4YY474 pKa = 10.45DD475 pKa = 3.87AKK477 pKa = 10.11VTTEE481 pKa = 4.16SGAEE485 pKa = 4.09VNNGTSNDD493 pKa = 3.22FGYY496 pKa = 10.91LYY498 pKa = 10.76LARR501 pKa = 11.84FHH503 pKa = 6.27MFPVTSVTSIPMLPSATLTTLGPLLSGSSLTEE535 pKa = 3.53YY536 pKa = 11.07ARR538 pKa = 11.84DD539 pKa = 3.5LRR541 pKa = 11.84LTTLSFNYY549 pKa = 10.33DD550 pKa = 4.28LIARR554 pKa = 11.84LWPMIADD561 pKa = 3.4NLEE564 pKa = 3.95NQTGYY569 pKa = 11.22SFIRR573 pKa = 11.84QIIVLLDD580 pKa = 3.85LLCDD584 pKa = 3.77LTGVIPGTEE593 pKa = 3.99YY594 pKa = 11.64GEE596 pKa = 4.28ITSNLYY602 pKa = 8.63VQPTARR608 pKa = 11.84HH609 pKa = 5.79RR610 pKa = 11.84NNEE613 pKa = 3.76NAAVFDD619 pKa = 4.27DD620 pKa = 4.28SGWNAFRR627 pKa = 11.84TEE629 pKa = 3.88RR630 pKa = 11.84VAQADD635 pKa = 3.83GRR637 pKa = 11.84WIKK640 pKa = 10.03FAILDD645 pKa = 3.7EE646 pKa = 4.27VTYY649 pKa = 10.86GKK651 pKa = 10.37VEE653 pKa = 4.04TGNFSTSDD661 pKa = 3.2FDD663 pKa = 6.16GIFTPTNWGKK673 pKa = 10.16NSAIVFVPIQKK684 pKa = 9.1TNDD687 pKa = 3.71FKK689 pKa = 11.12WIAAWTLAHH698 pKa = 7.4LAHH701 pKa = 6.94PWSNYY706 pKa = 8.89GATHH710 pKa = 6.93TYY712 pKa = 8.57ATTRR716 pKa = 11.84EE717 pKa = 4.15NGDD720 pKa = 3.09GYY722 pKa = 11.54AFHH725 pKa = 7.38TDD727 pKa = 3.37PNSNAIFNTNYY738 pKa = 9.83GCALAGPAMGVLYY751 pKa = 10.41VIVGTVKK758 pKa = 10.83LEE760 pKa = 3.7EE761 pKa = 3.93ARR763 pKa = 11.84INFAGRR769 pKa = 11.84VLTGDD774 pKa = 5.42DD775 pKa = 4.28IPPSEE780 pKa = 4.27PFEE783 pKa = 4.59FGDD786 pKa = 4.56NGFDD790 pKa = 3.42DD791 pKa = 5.15YY792 pKa = 11.47YY793 pKa = 11.74NVMVDD798 pKa = 3.3PSITAVCEE806 pKa = 4.45CIRR809 pKa = 11.84FWEE812 pKa = 4.58EE813 pKa = 3.75TFGTASEE820 pKa = 4.37RR821 pKa = 11.84ASAMRR826 pKa = 11.84IVASNRR832 pKa = 11.84KK833 pKa = 9.03CYY835 pKa = 9.97KK836 pKa = 10.35DD837 pKa = 3.5KK838 pKa = 11.61NFMGNGQLPGQVIASDD854 pKa = 4.08KK855 pKa = 10.95YY856 pKa = 9.42STHH859 pKa = 6.01QFATGPKK866 pKa = 8.04IWQGLGNGDD875 pKa = 3.22IANWSDD881 pKa = 3.54TVSSNDD887 pKa = 3.62LLTAQAVLAFGCQNALLMKK906 pKa = 9.89PVLRR910 pKa = 11.84GIRR913 pKa = 11.84SSSAAPVTGEE923 pKa = 4.84SEE925 pKa = 4.62TNLNTQLVFSEE936 pKa = 4.3RR937 pKa = 11.84NPLVEE942 pKa = 5.08LFVLKK947 pKa = 10.72KK948 pKa = 10.49LLTPAEE954 pKa = 4.4EE955 pKa = 4.57YY956 pKa = 10.33PVTRR960 pKa = 11.84LDD962 pKa = 3.26KK963 pKa = 10.54HH964 pKa = 5.88VRR966 pKa = 11.84TVTVLSAMCDD976 pKa = 3.49EE977 pKa = 4.46IAAMVDD983 pKa = 3.86LSCQANSVPSAIACYY998 pKa = 9.66PEE1000 pKa = 5.82LYY1002 pKa = 9.95TEE1004 pKa = 4.44SANICAFVRR1013 pKa = 11.84NKK1015 pKa = 10.05INPICSGILTLGIQFPQVFYY1035 pKa = 11.32ADD1037 pKa = 3.85TEE1039 pKa = 4.7SFVYY1043 pKa = 9.53TLEE1046 pKa = 4.08RR1047 pKa = 11.84ARR1049 pKa = 11.84LLSATINSDD1058 pKa = 3.13NYY1060 pKa = 10.5VNSNYY1065 pKa = 10.52LIGLSRR1071 pKa = 11.84IPTHH1075 pKa = 5.86TVSQYY1080 pKa = 10.78VPSLQDD1086 pKa = 3.04NRR1088 pKa = 11.84TGMQLSADD1096 pKa = 3.8VLRR1099 pKa = 11.84PRR1101 pKa = 11.84LGKK1104 pKa = 10.53FSGLVPNDD1112 pKa = 3.52WEE1114 pKa = 4.46GSTFFSVDD1122 pKa = 3.37SASDD1126 pKa = 3.52NMDD1129 pKa = 3.92DD1130 pKa = 6.17DD1131 pKa = 4.59ISIDD1135 pKa = 3.45NLLRR1139 pKa = 11.84PLSRR1143 pKa = 11.84GVIINSVNATSTNYY1157 pKa = 10.09GFAIASEE1164 pKa = 4.67GGNTAYY1170 pKa = 9.76PVLVHH1175 pKa = 5.74LTSWDD1180 pKa = 3.62FVINTIFTKK1189 pKa = 10.76AMNDD1193 pKa = 3.52PNITIPPGDD1202 pKa = 3.68SGMTPHH1208 pKa = 7.76DD1209 pKa = 3.71EE1210 pKa = 4.58RR1211 pKa = 11.84ILRR1214 pKa = 11.84RR1215 pKa = 11.84VPVDD1219 pKa = 2.84AWRR1222 pKa = 11.84RR1223 pKa = 11.84GRR1225 pKa = 11.84IIMTGPRR1232 pKa = 11.84DD1233 pKa = 3.66LFSPGSGPVYY1243 pKa = 10.22AYY1245 pKa = 11.27SLFSQVNSPSNIQTTKK1261 pKa = 10.3AWNADD1266 pKa = 2.94NDD1268 pKa = 4.19SAYY1271 pKa = 10.51RR1272 pKa = 11.84AADD1275 pKa = 3.41LGTVFKK1281 pKa = 10.26YY1282 pKa = 8.76TT1283 pKa = 3.41
MM1 pKa = 7.62AFLIPALVGGIEE13 pKa = 3.92AAVAGGGAVLGNKK26 pKa = 8.94LAHH29 pKa = 6.36KK30 pKa = 10.53VVGDD34 pKa = 4.18APADD38 pKa = 3.44LEE40 pKa = 4.76GIMEE44 pKa = 4.26VMQFLDD50 pKa = 3.8PSQNPSSMPRR60 pKa = 11.84MLQSQEE66 pKa = 4.28DD67 pKa = 3.94LSSALEE73 pKa = 3.94TRR75 pKa = 11.84LNIQGLDD82 pKa = 3.54NEE84 pKa = 4.76KK85 pKa = 9.24KK86 pKa = 8.89TKK88 pKa = 9.89FVSNLQALKK97 pKa = 10.57GQADD101 pKa = 4.16LALKK105 pKa = 10.56DD106 pKa = 3.82LLRR109 pKa = 11.84LKK111 pKa = 9.46STQGLSDD118 pKa = 3.29ATAVMNQAPNLIQNQPSLTSNPVPQAQVVTSPGLNVQARR157 pKa = 11.84EE158 pKa = 4.25APPTLQTLIQNLQAQAVPQSQVQDD182 pKa = 3.48SSALTRR188 pKa = 11.84NEE190 pKa = 3.66PSTTFSAVYY199 pKa = 9.34KK200 pKa = 9.87YY201 pKa = 10.1PVSRR205 pKa = 11.84LPLQASQPISKK216 pKa = 10.22NVLNQGTVIPRR227 pKa = 11.84PQVEE231 pKa = 4.69GTSSSQGTTNLRR243 pKa = 11.84TTASDD248 pKa = 3.52TVQGVGQALKK258 pKa = 10.68GAASDD263 pKa = 3.57AGKK266 pKa = 10.61VAVTALGNAAAEE278 pKa = 4.29VGKK281 pKa = 10.77GVLSHH286 pKa = 6.96LGKK289 pKa = 10.41LAAAKK294 pKa = 10.24IGDD297 pKa = 3.99FSSILAKK304 pKa = 10.48KK305 pKa = 9.95IVGDD309 pKa = 3.8AVPANDD315 pKa = 3.92HH316 pKa = 7.47RR317 pKa = 11.84IGTWMRR323 pKa = 11.84SEE325 pKa = 4.24KK326 pKa = 10.7VVGDD330 pKa = 4.06AEE332 pKa = 4.79VSTKK336 pKa = 10.94DD337 pKa = 3.55NPKK340 pKa = 10.7LMALLQQMANGGSAPKK356 pKa = 10.37SGEE359 pKa = 3.95DD360 pKa = 3.27KK361 pKa = 10.88TDD363 pKa = 3.15GKK365 pKa = 10.7IVGDD369 pKa = 3.91APPGVMEE376 pKa = 4.71KK377 pKa = 10.17QDD379 pKa = 3.4EE380 pKa = 4.68TMTDD384 pKa = 3.39EE385 pKa = 4.35QVAAAAGNLQSSPSLALMSHH405 pKa = 6.31IQLDD409 pKa = 4.04EE410 pKa = 4.03QMLEE414 pKa = 3.91SGKK417 pKa = 10.24VVEE420 pKa = 4.86FTSDD424 pKa = 2.87VSEE427 pKa = 4.33AARR430 pKa = 11.84MIARR434 pKa = 11.84LGEE437 pKa = 4.12EE438 pKa = 3.88VSGVFKK444 pKa = 9.84TEE446 pKa = 3.66RR447 pKa = 11.84LISTRR452 pKa = 11.84TQVGNQYY459 pKa = 10.05FWNTYY464 pKa = 6.58FTLRR468 pKa = 11.84NPRR471 pKa = 11.84LDD473 pKa = 3.4YY474 pKa = 10.45DD475 pKa = 3.87AKK477 pKa = 10.11VTTEE481 pKa = 4.16SGAEE485 pKa = 4.09VNNGTSNDD493 pKa = 3.22FGYY496 pKa = 10.91LYY498 pKa = 10.76LARR501 pKa = 11.84FHH503 pKa = 6.27MFPVTSVTSIPMLPSATLTTLGPLLSGSSLTEE535 pKa = 3.53YY536 pKa = 11.07ARR538 pKa = 11.84DD539 pKa = 3.5LRR541 pKa = 11.84LTTLSFNYY549 pKa = 10.33DD550 pKa = 4.28LIARR554 pKa = 11.84LWPMIADD561 pKa = 3.4NLEE564 pKa = 3.95NQTGYY569 pKa = 11.22SFIRR573 pKa = 11.84QIIVLLDD580 pKa = 3.85LLCDD584 pKa = 3.77LTGVIPGTEE593 pKa = 3.99YY594 pKa = 11.64GEE596 pKa = 4.28ITSNLYY602 pKa = 8.63VQPTARR608 pKa = 11.84HH609 pKa = 5.79RR610 pKa = 11.84NNEE613 pKa = 3.76NAAVFDD619 pKa = 4.27DD620 pKa = 4.28SGWNAFRR627 pKa = 11.84TEE629 pKa = 3.88RR630 pKa = 11.84VAQADD635 pKa = 3.83GRR637 pKa = 11.84WIKK640 pKa = 10.03FAILDD645 pKa = 3.7EE646 pKa = 4.27VTYY649 pKa = 10.86GKK651 pKa = 10.37VEE653 pKa = 4.04TGNFSTSDD661 pKa = 3.2FDD663 pKa = 6.16GIFTPTNWGKK673 pKa = 10.16NSAIVFVPIQKK684 pKa = 9.1TNDD687 pKa = 3.71FKK689 pKa = 11.12WIAAWTLAHH698 pKa = 7.4LAHH701 pKa = 6.94PWSNYY706 pKa = 8.89GATHH710 pKa = 6.93TYY712 pKa = 8.57ATTRR716 pKa = 11.84EE717 pKa = 4.15NGDD720 pKa = 3.09GYY722 pKa = 11.54AFHH725 pKa = 7.38TDD727 pKa = 3.37PNSNAIFNTNYY738 pKa = 9.83GCALAGPAMGVLYY751 pKa = 10.41VIVGTVKK758 pKa = 10.83LEE760 pKa = 3.7EE761 pKa = 3.93ARR763 pKa = 11.84INFAGRR769 pKa = 11.84VLTGDD774 pKa = 5.42DD775 pKa = 4.28IPPSEE780 pKa = 4.27PFEE783 pKa = 4.59FGDD786 pKa = 4.56NGFDD790 pKa = 3.42DD791 pKa = 5.15YY792 pKa = 11.47YY793 pKa = 11.74NVMVDD798 pKa = 3.3PSITAVCEE806 pKa = 4.45CIRR809 pKa = 11.84FWEE812 pKa = 4.58EE813 pKa = 3.75TFGTASEE820 pKa = 4.37RR821 pKa = 11.84ASAMRR826 pKa = 11.84IVASNRR832 pKa = 11.84KK833 pKa = 9.03CYY835 pKa = 9.97KK836 pKa = 10.35DD837 pKa = 3.5KK838 pKa = 11.61NFMGNGQLPGQVIASDD854 pKa = 4.08KK855 pKa = 10.95YY856 pKa = 9.42STHH859 pKa = 6.01QFATGPKK866 pKa = 8.04IWQGLGNGDD875 pKa = 3.22IANWSDD881 pKa = 3.54TVSSNDD887 pKa = 3.62LLTAQAVLAFGCQNALLMKK906 pKa = 9.89PVLRR910 pKa = 11.84GIRR913 pKa = 11.84SSSAAPVTGEE923 pKa = 4.84SEE925 pKa = 4.62TNLNTQLVFSEE936 pKa = 4.3RR937 pKa = 11.84NPLVEE942 pKa = 5.08LFVLKK947 pKa = 10.72KK948 pKa = 10.49LLTPAEE954 pKa = 4.4EE955 pKa = 4.57YY956 pKa = 10.33PVTRR960 pKa = 11.84LDD962 pKa = 3.26KK963 pKa = 10.54HH964 pKa = 5.88VRR966 pKa = 11.84TVTVLSAMCDD976 pKa = 3.49EE977 pKa = 4.46IAAMVDD983 pKa = 3.86LSCQANSVPSAIACYY998 pKa = 9.66PEE1000 pKa = 5.82LYY1002 pKa = 9.95TEE1004 pKa = 4.44SANICAFVRR1013 pKa = 11.84NKK1015 pKa = 10.05INPICSGILTLGIQFPQVFYY1035 pKa = 11.32ADD1037 pKa = 3.85TEE1039 pKa = 4.7SFVYY1043 pKa = 9.53TLEE1046 pKa = 4.08RR1047 pKa = 11.84ARR1049 pKa = 11.84LLSATINSDD1058 pKa = 3.13NYY1060 pKa = 10.5VNSNYY1065 pKa = 10.52LIGLSRR1071 pKa = 11.84IPTHH1075 pKa = 5.86TVSQYY1080 pKa = 10.78VPSLQDD1086 pKa = 3.04NRR1088 pKa = 11.84TGMQLSADD1096 pKa = 3.8VLRR1099 pKa = 11.84PRR1101 pKa = 11.84LGKK1104 pKa = 10.53FSGLVPNDD1112 pKa = 3.52WEE1114 pKa = 4.46GSTFFSVDD1122 pKa = 3.37SASDD1126 pKa = 3.52NMDD1129 pKa = 3.92DD1130 pKa = 6.17DD1131 pKa = 4.59ISIDD1135 pKa = 3.45NLLRR1139 pKa = 11.84PLSRR1143 pKa = 11.84GVIINSVNATSTNYY1157 pKa = 10.09GFAIASEE1164 pKa = 4.67GGNTAYY1170 pKa = 9.76PVLVHH1175 pKa = 5.74LTSWDD1180 pKa = 3.62FVINTIFTKK1189 pKa = 10.76AMNDD1193 pKa = 3.52PNITIPPGDD1202 pKa = 3.68SGMTPHH1208 pKa = 7.76DD1209 pKa = 3.71EE1210 pKa = 4.58RR1211 pKa = 11.84ILRR1214 pKa = 11.84RR1215 pKa = 11.84VPVDD1219 pKa = 2.84AWRR1222 pKa = 11.84RR1223 pKa = 11.84GRR1225 pKa = 11.84IIMTGPRR1232 pKa = 11.84DD1233 pKa = 3.66LFSPGSGPVYY1243 pKa = 10.22AYY1245 pKa = 11.27SLFSQVNSPSNIQTTKK1261 pKa = 10.3AWNADD1266 pKa = 2.94NDD1268 pKa = 4.19SAYY1271 pKa = 10.51RR1272 pKa = 11.84AADD1275 pKa = 3.41LGTVFKK1281 pKa = 10.26YY1282 pKa = 8.76TT1283 pKa = 3.41
Molecular weight: 138.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF62|A0A1L3KF62_9VIRU Uncharacterized protein OS=Beihai sea slater virus 3 OX=1922659 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 4.99GFFNPSSSRR11 pKa = 11.84GHH13 pKa = 6.38RR14 pKa = 11.84GGSGRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84CSAWKK26 pKa = 9.35QARR29 pKa = 11.84TQSRR33 pKa = 11.84RR34 pKa = 11.84GRR36 pKa = 11.84ASRR39 pKa = 11.84PGRR42 pKa = 11.84DD43 pKa = 3.38NGSDD47 pKa = 3.42AILRR51 pKa = 11.84PKK53 pKa = 10.52SKK55 pKa = 10.45SFKK58 pKa = 9.2HH59 pKa = 5.96ASNASIPRR67 pKa = 11.84RR68 pKa = 11.84LEE70 pKa = 3.33QCIRR74 pKa = 11.84NTIEE78 pKa = 3.59HH79 pKa = 6.15SRR81 pKa = 11.84FGQRR85 pKa = 11.84KK86 pKa = 6.74EE87 pKa = 3.76NKK89 pKa = 9.24IRR91 pKa = 11.84FQPAGVEE98 pKa = 4.41GASGLGVEE106 pKa = 4.99RR107 pKa = 11.84LTQAEE112 pKa = 3.52IDD114 pKa = 3.69ARR116 pKa = 11.84AEE118 pKa = 3.63RR119 pKa = 11.84CYY121 pKa = 10.99CRR123 pKa = 11.84DD124 pKa = 3.3EE125 pKa = 4.94SGTEE129 pKa = 3.99SDD131 pKa = 4.55SEE133 pKa = 3.84PAIIDD138 pKa = 3.98FEE140 pKa = 4.58PSSPGTGGDD149 pKa = 3.08IPGFEE154 pKa = 4.62CAGEE158 pKa = 3.98RR159 pKa = 11.84SSPNPPNIDD168 pKa = 3.79PEE170 pKa = 4.44PTSSGSATEE179 pKa = 4.13PSSGQLSPNTKK190 pKa = 9.76RR191 pKa = 11.84ALDD194 pKa = 3.78NLLSCIQVSSFPPSTASLAAYY215 pKa = 9.94LKK217 pKa = 10.11EE218 pKa = 4.24CVEE221 pKa = 4.46SRR223 pKa = 11.84DD224 pKa = 4.02SNTKK228 pKa = 10.29ASSS231 pKa = 3.0
MM1 pKa = 8.1DD2 pKa = 4.99GFFNPSSSRR11 pKa = 11.84GHH13 pKa = 6.38RR14 pKa = 11.84GGSGRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84CSAWKK26 pKa = 9.35QARR29 pKa = 11.84TQSRR33 pKa = 11.84RR34 pKa = 11.84GRR36 pKa = 11.84ASRR39 pKa = 11.84PGRR42 pKa = 11.84DD43 pKa = 3.38NGSDD47 pKa = 3.42AILRR51 pKa = 11.84PKK53 pKa = 10.52SKK55 pKa = 10.45SFKK58 pKa = 9.2HH59 pKa = 5.96ASNASIPRR67 pKa = 11.84RR68 pKa = 11.84LEE70 pKa = 3.33QCIRR74 pKa = 11.84NTIEE78 pKa = 3.59HH79 pKa = 6.15SRR81 pKa = 11.84FGQRR85 pKa = 11.84KK86 pKa = 6.74EE87 pKa = 3.76NKK89 pKa = 9.24IRR91 pKa = 11.84FQPAGVEE98 pKa = 4.41GASGLGVEE106 pKa = 4.99RR107 pKa = 11.84LTQAEE112 pKa = 3.52IDD114 pKa = 3.69ARR116 pKa = 11.84AEE118 pKa = 3.63RR119 pKa = 11.84CYY121 pKa = 10.99CRR123 pKa = 11.84DD124 pKa = 3.3EE125 pKa = 4.94SGTEE129 pKa = 3.99SDD131 pKa = 4.55SEE133 pKa = 3.84PAIIDD138 pKa = 3.98FEE140 pKa = 4.58PSSPGTGGDD149 pKa = 3.08IPGFEE154 pKa = 4.62CAGEE158 pKa = 3.98RR159 pKa = 11.84SSPNPPNIDD168 pKa = 3.79PEE170 pKa = 4.44PTSSGSATEE179 pKa = 4.13PSSGQLSPNTKK190 pKa = 9.76RR191 pKa = 11.84ALDD194 pKa = 3.78NLLSCIQVSSFPPSTASLAAYY215 pKa = 9.94LKK217 pKa = 10.11EE218 pKa = 4.24CVEE221 pKa = 4.46SRR223 pKa = 11.84DD224 pKa = 4.02SNTKK228 pKa = 10.29ASSS231 pKa = 3.0
Molecular weight: 24.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2377 |
231 |
1283 |
792.3 |
88.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.615 ± 1.487 | 1.346 ± 0.452 |
6.31 ± 0.621 | 4.964 ± 0.599 |
4.712 ± 0.89 | 6.353 ± 1.056 |
1.388 ± 0.181 | 5.721 ± 0.674 |
5.385 ± 1.495 | 8.961 ± 1.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.893 ± 0.366 | 5.974 ± 0.361 |
4.838 ± 0.832 | 3.45 ± 0.79 |
5.595 ± 1.308 | 7.993 ± 1.739 |
6.899 ± 0.687 | 6.353 ± 1.084 |
1.388 ± 0.265 | 2.861 ± 0.452 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
