
Rodent Torque teno virus 3
Taxonomy: Viruses; Anelloviridae; Wawtorquevirus; Torque teno rodent virus 2
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
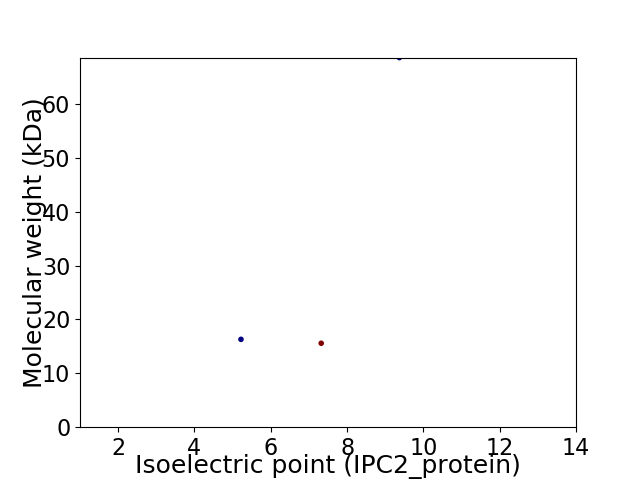
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
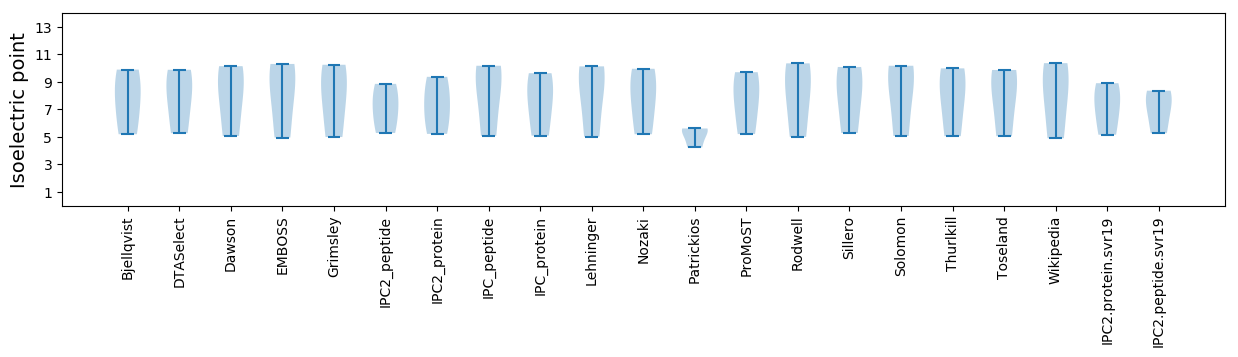
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4QBE6|A0A2H4QBE6_9VIRU Uncharacterized protein OS=Rodent Torque teno virus 3 OX=2054610 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.02FSAAAKK8 pKa = 9.5ILLTIFLDD16 pKa = 4.68GVLLPALGRR25 pKa = 11.84PSPLWLQEE33 pKa = 3.99LEE35 pKa = 4.34KK36 pKa = 10.54ATVVPEE42 pKa = 4.03TVEE45 pKa = 4.31NKK47 pKa = 7.29TTPLPGKK54 pKa = 7.35EE55 pKa = 4.06TPQAVVEE62 pKa = 4.55TEE64 pKa = 3.95NQACTCHH71 pKa = 7.32AMEE74 pKa = 5.76PDD76 pKa = 3.45QQKK79 pKa = 10.54EE80 pKa = 4.45MYY82 pKa = 9.61HH83 pKa = 6.36SWILSFSILLGWPEE97 pKa = 3.97GDD99 pKa = 3.99NGFHH103 pKa = 7.48LPRR106 pKa = 11.84EE107 pKa = 4.36QADD110 pKa = 2.69SWMARR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84CSQQPSVSPRR127 pKa = 11.84PVLGGEE133 pKa = 4.2VLEE136 pKa = 4.63STVEE140 pKa = 4.04QQQSRR145 pKa = 11.84LL146 pKa = 3.56
MM1 pKa = 7.41KK2 pKa = 10.02FSAAAKK8 pKa = 9.5ILLTIFLDD16 pKa = 4.68GVLLPALGRR25 pKa = 11.84PSPLWLQEE33 pKa = 3.99LEE35 pKa = 4.34KK36 pKa = 10.54ATVVPEE42 pKa = 4.03TVEE45 pKa = 4.31NKK47 pKa = 7.29TTPLPGKK54 pKa = 7.35EE55 pKa = 4.06TPQAVVEE62 pKa = 4.55TEE64 pKa = 3.95NQACTCHH71 pKa = 7.32AMEE74 pKa = 5.76PDD76 pKa = 3.45QQKK79 pKa = 10.54EE80 pKa = 4.45MYY82 pKa = 9.61HH83 pKa = 6.36SWILSFSILLGWPEE97 pKa = 3.97GDD99 pKa = 3.99NGFHH103 pKa = 7.48LPRR106 pKa = 11.84EE107 pKa = 4.36QADD110 pKa = 2.69SWMARR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84CSQQPSVSPRR127 pKa = 11.84PVLGGEE133 pKa = 4.2VLEE136 pKa = 4.63STVEE140 pKa = 4.04QQQSRR145 pKa = 11.84LL146 pKa = 3.56
Molecular weight: 16.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4QBE5|A0A2H4QBE5_9VIRU Uncharacterized protein OS=Rodent Torque teno virus 3 OX=2054610 PE=4 SV=1
MM1 pKa = 7.58AFYY4 pKa = 10.63SRR6 pKa = 11.84RR7 pKa = 11.84WGGLRR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.84GYY16 pKa = 10.04RR17 pKa = 11.84NWRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84PWYY25 pKa = 8.56RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84WRR30 pKa = 11.84TRR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 8.76RR37 pKa = 11.84VRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84LLRR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 7.1TRR51 pKa = 11.84HH52 pKa = 5.01VPVMQWNPINKK63 pKa = 8.79KK64 pKa = 9.04RR65 pKa = 11.84CTIRR69 pKa = 11.84GFCPLVYY76 pKa = 10.47CLGGQKK82 pKa = 10.81GIMDD86 pKa = 3.89FTYY89 pKa = 10.34PGNKK93 pKa = 9.73LILGWLGGGVHH104 pKa = 6.77SSLLSLLDD112 pKa = 3.87LFWEE116 pKa = 4.1EE117 pKa = 4.41RR118 pKa = 11.84YY119 pKa = 8.89WRR121 pKa = 11.84ARR123 pKa = 11.84WSSSNQGYY131 pKa = 9.16NLFRR135 pKa = 11.84YY136 pKa = 9.44YY137 pKa = 11.08GATITLYY144 pKa = 9.2PVMYY148 pKa = 8.1YY149 pKa = 9.9TYY151 pKa = 9.61IFWYY155 pKa = 8.2STEE158 pKa = 3.97EE159 pKa = 3.8LTEE162 pKa = 4.16DD163 pKa = 3.79VEE165 pKa = 4.56PLTVCHH171 pKa = 6.78PSQLLLSKK179 pKa = 10.23HH180 pKa = 5.53HH181 pKa = 6.56VIVHH185 pKa = 6.5RR186 pKa = 11.84LDD188 pKa = 4.4GFQKK192 pKa = 8.88TKK194 pKa = 10.21PIKK197 pKa = 10.47LHH199 pKa = 6.28IKK201 pKa = 9.84PPSQMTGTWHH211 pKa = 6.94TFAAWAKK218 pKa = 10.07QPLLKK223 pKa = 10.06WRR225 pKa = 11.84ISLLDD230 pKa = 3.97IEE232 pKa = 5.05NPWTGFPNDD241 pKa = 3.41QTQGVHH247 pKa = 4.23CTVWARR253 pKa = 11.84HH254 pKa = 4.04KK255 pKa = 9.27TEE257 pKa = 4.4RR258 pKa = 11.84PTEE261 pKa = 4.0ARR263 pKa = 11.84SLDD266 pKa = 3.37LWYY269 pKa = 10.56FPLLDD274 pKa = 5.34DD275 pKa = 4.6GTDD278 pKa = 3.64LSVTMHH284 pKa = 6.9KK285 pKa = 10.39IKK287 pKa = 10.14WNNDD291 pKa = 2.6GTGPKK296 pKa = 9.73YY297 pKa = 10.77DD298 pKa = 4.37EE299 pKa = 4.69ANFWPVAAQFQRR311 pKa = 11.84LLVPFYY317 pKa = 9.61MYY319 pKa = 11.21GFGRR323 pKa = 11.84SATFYY328 pKa = 11.52DD329 pKa = 4.12MNEE332 pKa = 4.29TTHH335 pKa = 6.57EE336 pKa = 4.48PGPTEE341 pKa = 4.06GNMGSFLFLKK351 pKa = 10.12FLNPPCWRR359 pKa = 11.84GSEE362 pKa = 4.25GQFPQFKK369 pKa = 10.66DD370 pKa = 3.64DD371 pKa = 3.61MCFMTFGTVAQLCAQGPWCMKK392 pKa = 10.46RR393 pKa = 11.84IGTNTAGVNISMKK406 pKa = 10.38YY407 pKa = 10.19KK408 pKa = 10.59FYY410 pKa = 10.14FQWGGTPGTNLPPVQPVAGNPGQPLLQSTLRR441 pKa = 11.84WGNTIRR447 pKa = 11.84ADD449 pKa = 3.16IRR451 pKa = 11.84DD452 pKa = 3.86PTTVGEE458 pKa = 4.12EE459 pKa = 4.28VLWPEE464 pKa = 4.96DD465 pKa = 3.83LDD467 pKa = 3.48EE468 pKa = 6.09HH469 pKa = 7.45GIITSQAFRR478 pKa = 11.84RR479 pKa = 11.84ITQPSVSTGSRR490 pKa = 11.84SLGTLGCLRR499 pKa = 11.84PAVSSGRR506 pKa = 11.84KK507 pKa = 8.68RR508 pKa = 11.84PRR510 pKa = 11.84HH511 pKa = 4.95SFEE514 pKa = 3.77EE515 pKa = 4.51SEE517 pKa = 4.4EE518 pKa = 4.32EE519 pKa = 4.08EE520 pKa = 4.62HH521 pKa = 7.49SSSPQEE527 pKa = 3.91TEE529 pKa = 3.79NYY531 pKa = 8.14STEE534 pKa = 4.11EE535 pKa = 4.02EE536 pKa = 4.24EE537 pKa = 5.04EE538 pKa = 3.96EE539 pKa = 4.02MEE541 pKa = 3.96RR542 pKa = 11.84ARR544 pKa = 11.84RR545 pKa = 11.84KK546 pKa = 5.89QHH548 pKa = 4.17RR549 pKa = 11.84RR550 pKa = 11.84KK551 pKa = 10.0RR552 pKa = 11.84KK553 pKa = 9.48RR554 pKa = 11.84LEE556 pKa = 3.47QLLRR560 pKa = 11.84GLSNLGFRR568 pKa = 11.84RR569 pKa = 11.84PMMAGPSGNYY579 pKa = 8.88QKK581 pKa = 11.25SNSII585 pKa = 3.71
MM1 pKa = 7.58AFYY4 pKa = 10.63SRR6 pKa = 11.84RR7 pKa = 11.84WGGLRR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.84GYY16 pKa = 10.04RR17 pKa = 11.84NWRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84PWYY25 pKa = 8.56RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84WRR30 pKa = 11.84TRR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 8.76RR37 pKa = 11.84VRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84LLRR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 7.1TRR51 pKa = 11.84HH52 pKa = 5.01VPVMQWNPINKK63 pKa = 8.79KK64 pKa = 9.04RR65 pKa = 11.84CTIRR69 pKa = 11.84GFCPLVYY76 pKa = 10.47CLGGQKK82 pKa = 10.81GIMDD86 pKa = 3.89FTYY89 pKa = 10.34PGNKK93 pKa = 9.73LILGWLGGGVHH104 pKa = 6.77SSLLSLLDD112 pKa = 3.87LFWEE116 pKa = 4.1EE117 pKa = 4.41RR118 pKa = 11.84YY119 pKa = 8.89WRR121 pKa = 11.84ARR123 pKa = 11.84WSSSNQGYY131 pKa = 9.16NLFRR135 pKa = 11.84YY136 pKa = 9.44YY137 pKa = 11.08GATITLYY144 pKa = 9.2PVMYY148 pKa = 8.1YY149 pKa = 9.9TYY151 pKa = 9.61IFWYY155 pKa = 8.2STEE158 pKa = 3.97EE159 pKa = 3.8LTEE162 pKa = 4.16DD163 pKa = 3.79VEE165 pKa = 4.56PLTVCHH171 pKa = 6.78PSQLLLSKK179 pKa = 10.23HH180 pKa = 5.53HH181 pKa = 6.56VIVHH185 pKa = 6.5RR186 pKa = 11.84LDD188 pKa = 4.4GFQKK192 pKa = 8.88TKK194 pKa = 10.21PIKK197 pKa = 10.47LHH199 pKa = 6.28IKK201 pKa = 9.84PPSQMTGTWHH211 pKa = 6.94TFAAWAKK218 pKa = 10.07QPLLKK223 pKa = 10.06WRR225 pKa = 11.84ISLLDD230 pKa = 3.97IEE232 pKa = 5.05NPWTGFPNDD241 pKa = 3.41QTQGVHH247 pKa = 4.23CTVWARR253 pKa = 11.84HH254 pKa = 4.04KK255 pKa = 9.27TEE257 pKa = 4.4RR258 pKa = 11.84PTEE261 pKa = 4.0ARR263 pKa = 11.84SLDD266 pKa = 3.37LWYY269 pKa = 10.56FPLLDD274 pKa = 5.34DD275 pKa = 4.6GTDD278 pKa = 3.64LSVTMHH284 pKa = 6.9KK285 pKa = 10.39IKK287 pKa = 10.14WNNDD291 pKa = 2.6GTGPKK296 pKa = 9.73YY297 pKa = 10.77DD298 pKa = 4.37EE299 pKa = 4.69ANFWPVAAQFQRR311 pKa = 11.84LLVPFYY317 pKa = 9.61MYY319 pKa = 11.21GFGRR323 pKa = 11.84SATFYY328 pKa = 11.52DD329 pKa = 4.12MNEE332 pKa = 4.29TTHH335 pKa = 6.57EE336 pKa = 4.48PGPTEE341 pKa = 4.06GNMGSFLFLKK351 pKa = 10.12FLNPPCWRR359 pKa = 11.84GSEE362 pKa = 4.25GQFPQFKK369 pKa = 10.66DD370 pKa = 3.64DD371 pKa = 3.61MCFMTFGTVAQLCAQGPWCMKK392 pKa = 10.46RR393 pKa = 11.84IGTNTAGVNISMKK406 pKa = 10.38YY407 pKa = 10.19KK408 pKa = 10.59FYY410 pKa = 10.14FQWGGTPGTNLPPVQPVAGNPGQPLLQSTLRR441 pKa = 11.84WGNTIRR447 pKa = 11.84ADD449 pKa = 3.16IRR451 pKa = 11.84DD452 pKa = 3.86PTTVGEE458 pKa = 4.12EE459 pKa = 4.28VLWPEE464 pKa = 4.96DD465 pKa = 3.83LDD467 pKa = 3.48EE468 pKa = 6.09HH469 pKa = 7.45GIITSQAFRR478 pKa = 11.84RR479 pKa = 11.84ITQPSVSTGSRR490 pKa = 11.84SLGTLGCLRR499 pKa = 11.84PAVSSGRR506 pKa = 11.84KK507 pKa = 8.68RR508 pKa = 11.84PRR510 pKa = 11.84HH511 pKa = 4.95SFEE514 pKa = 3.77EE515 pKa = 4.51SEE517 pKa = 4.4EE518 pKa = 4.32EE519 pKa = 4.08EE520 pKa = 4.62HH521 pKa = 7.49SSSPQEE527 pKa = 3.91TEE529 pKa = 3.79NYY531 pKa = 8.14STEE534 pKa = 4.11EE535 pKa = 4.02EE536 pKa = 4.24EE537 pKa = 5.04EE538 pKa = 3.96EE539 pKa = 4.02MEE541 pKa = 3.96RR542 pKa = 11.84ARR544 pKa = 11.84RR545 pKa = 11.84KK546 pKa = 5.89QHH548 pKa = 4.17RR549 pKa = 11.84RR550 pKa = 11.84KK551 pKa = 10.0RR552 pKa = 11.84KK553 pKa = 9.48RR554 pKa = 11.84LEE556 pKa = 3.47QLLRR560 pKa = 11.84GLSNLGFRR568 pKa = 11.84RR569 pKa = 11.84PMMAGPSGNYY579 pKa = 8.88QKK581 pKa = 11.25SNSII585 pKa = 3.71
Molecular weight: 68.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
871 |
140 |
585 |
290.3 |
33.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.396 ± 1.922 | 1.493 ± 0.405 |
3.674 ± 0.741 | 6.774 ± 1.15 |
4.133 ± 0.573 | 7.577 ± 0.805 |
2.411 ± 0.339 | 3.444 ± 0.289 |
4.822 ± 0.621 | 9.07 ± 0.886 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.526 ± 0.293 | 3.33 ± 0.647 |
7.348 ± 0.623 | 4.937 ± 0.945 |
8.955 ± 1.628 | 7.003 ± 0.919 |
6.544 ± 0.689 | 3.789 ± 1.274 |
3.559 ± 0.404 | 3.215 ± 1.172 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
