
Arctopus echinatus-associated virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
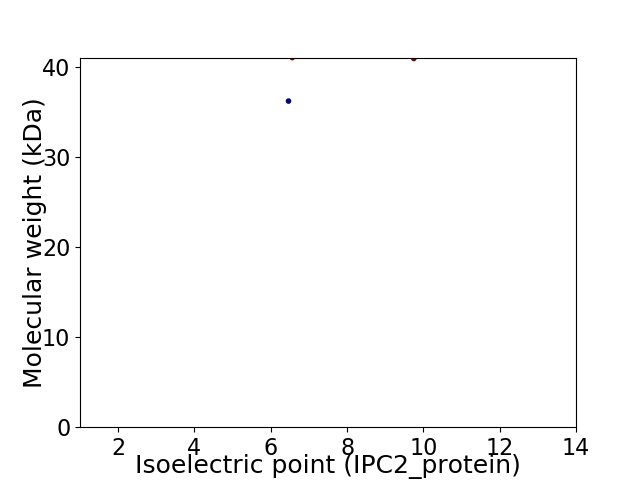
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
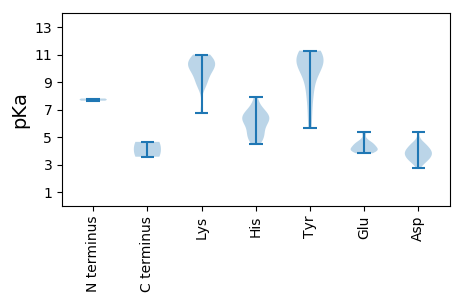
Protein with the lowest isoelectric point:
>tr|A0A345BKA2|A0A345BKA2_9VIRU Replication-associated protein OS=Arctopus echinatus-associated virus OX=2282642 PE=3 SV=1
MM1 pKa = 7.64SSGSTNQLLTLRR13 pKa = 11.84GRR15 pKa = 11.84YY16 pKa = 8.63FLLTYY21 pKa = 9.86PRR23 pKa = 11.84SDD25 pKa = 3.67FNHH28 pKa = 4.73AHH30 pKa = 6.59AWIQLNEE37 pKa = 4.11RR38 pKa = 11.84GACGAIVCSEE48 pKa = 3.91LHH50 pKa = 6.27QDD52 pKa = 3.78DD53 pKa = 4.24TEE55 pKa = 4.22HH56 pKa = 6.72RR57 pKa = 11.84HH58 pKa = 5.07IAVDD62 pKa = 4.1FGRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84QFSNAHH73 pKa = 5.01HH74 pKa = 6.65TFNITLGDD82 pKa = 3.89TTWTGNYY89 pKa = 5.64QTARR93 pKa = 11.84SWAAIRR99 pKa = 11.84HH100 pKa = 4.5YY101 pKa = 10.42VEE103 pKa = 4.45KK104 pKa = 10.44DD105 pKa = 3.23GNVSYY110 pKa = 10.85FGTSEE115 pKa = 4.2PNLEE119 pKa = 4.08QANPEE124 pKa = 4.28TEE126 pKa = 4.07TEE128 pKa = 4.45DD129 pKa = 4.64IIDD132 pKa = 3.94ALTRR136 pKa = 11.84TQSYY140 pKa = 10.91AEE142 pKa = 3.97WLVWTIKK149 pKa = 10.51QKK151 pKa = 10.76ISFGHH156 pKa = 7.1ANALWQEE163 pKa = 4.01LRR165 pKa = 11.84GNRR168 pKa = 11.84PATLLEE174 pKa = 4.32GDD176 pKa = 4.53NIPGNVVLPEE186 pKa = 3.91LQLRR190 pKa = 11.84TFSQDD195 pKa = 2.72TNRR198 pKa = 11.84ALVLLGPSGVGKK210 pKa = 6.77TTWAKK215 pKa = 11.0LNIPKK220 pKa = 9.67PALLVSHH227 pKa = 7.25IDD229 pKa = 3.54DD230 pKa = 4.2LKK232 pKa = 9.05HH233 pKa = 6.31FKK235 pKa = 10.93VGVHH239 pKa = 6.02KK240 pKa = 10.85GIIFDD245 pKa = 4.34DD246 pKa = 3.43MHH248 pKa = 6.75FAGDD252 pKa = 3.97EE253 pKa = 4.12NGKK256 pKa = 9.54GAWPRR261 pKa = 11.84TSQIHH266 pKa = 6.24LVDD269 pKa = 4.46FEE271 pKa = 4.46NPRR274 pKa = 11.84SIHH277 pKa = 5.65CRR279 pKa = 11.84HH280 pKa = 6.28RR281 pKa = 11.84CATIPAGVFKK291 pKa = 10.83IFTCNVYY298 pKa = 10.25PFSVDD303 pKa = 3.15PAIEE307 pKa = 3.82RR308 pKa = 11.84RR309 pKa = 11.84IYY311 pKa = 10.83LITLNFLQQ319 pKa = 4.65
MM1 pKa = 7.64SSGSTNQLLTLRR13 pKa = 11.84GRR15 pKa = 11.84YY16 pKa = 8.63FLLTYY21 pKa = 9.86PRR23 pKa = 11.84SDD25 pKa = 3.67FNHH28 pKa = 4.73AHH30 pKa = 6.59AWIQLNEE37 pKa = 4.11RR38 pKa = 11.84GACGAIVCSEE48 pKa = 3.91LHH50 pKa = 6.27QDD52 pKa = 3.78DD53 pKa = 4.24TEE55 pKa = 4.22HH56 pKa = 6.72RR57 pKa = 11.84HH58 pKa = 5.07IAVDD62 pKa = 4.1FGRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84QFSNAHH73 pKa = 5.01HH74 pKa = 6.65TFNITLGDD82 pKa = 3.89TTWTGNYY89 pKa = 5.64QTARR93 pKa = 11.84SWAAIRR99 pKa = 11.84HH100 pKa = 4.5YY101 pKa = 10.42VEE103 pKa = 4.45KK104 pKa = 10.44DD105 pKa = 3.23GNVSYY110 pKa = 10.85FGTSEE115 pKa = 4.2PNLEE119 pKa = 4.08QANPEE124 pKa = 4.28TEE126 pKa = 4.07TEE128 pKa = 4.45DD129 pKa = 4.64IIDD132 pKa = 3.94ALTRR136 pKa = 11.84TQSYY140 pKa = 10.91AEE142 pKa = 3.97WLVWTIKK149 pKa = 10.51QKK151 pKa = 10.76ISFGHH156 pKa = 7.1ANALWQEE163 pKa = 4.01LRR165 pKa = 11.84GNRR168 pKa = 11.84PATLLEE174 pKa = 4.32GDD176 pKa = 4.53NIPGNVVLPEE186 pKa = 3.91LQLRR190 pKa = 11.84TFSQDD195 pKa = 2.72TNRR198 pKa = 11.84ALVLLGPSGVGKK210 pKa = 6.77TTWAKK215 pKa = 11.0LNIPKK220 pKa = 9.67PALLVSHH227 pKa = 7.25IDD229 pKa = 3.54DD230 pKa = 4.2LKK232 pKa = 9.05HH233 pKa = 6.31FKK235 pKa = 10.93VGVHH239 pKa = 6.02KK240 pKa = 10.85GIIFDD245 pKa = 4.34DD246 pKa = 3.43MHH248 pKa = 6.75FAGDD252 pKa = 3.97EE253 pKa = 4.12NGKK256 pKa = 9.54GAWPRR261 pKa = 11.84TSQIHH266 pKa = 6.24LVDD269 pKa = 4.46FEE271 pKa = 4.46NPRR274 pKa = 11.84SIHH277 pKa = 5.65CRR279 pKa = 11.84HH280 pKa = 6.28RR281 pKa = 11.84CATIPAGVFKK291 pKa = 10.83IFTCNVYY298 pKa = 10.25PFSVDD303 pKa = 3.15PAIEE307 pKa = 3.82RR308 pKa = 11.84RR309 pKa = 11.84IYY311 pKa = 10.83LITLNFLQQ319 pKa = 4.65
Molecular weight: 36.19 kDa
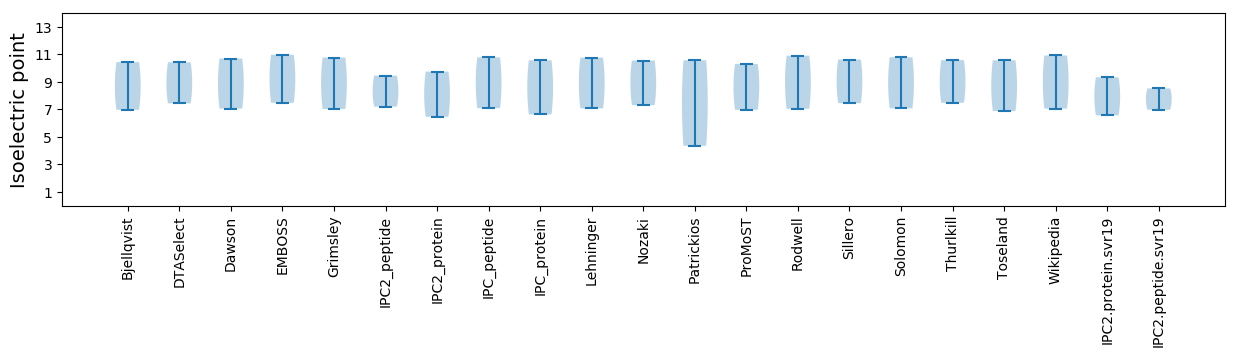
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BKA2|A0A345BKA2_9VIRU Replication-associated protein OS=Arctopus echinatus-associated virus OX=2282642 PE=3 SV=1
MM1 pKa = 7.8AKK3 pKa = 10.03RR4 pKa = 11.84SRR6 pKa = 11.84SRR8 pKa = 11.84SVSRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 10.19LPSNWHH22 pKa = 5.35MASNVARR29 pKa = 11.84AAGTAYY35 pKa = 10.42DD36 pKa = 3.32AYY38 pKa = 10.07KK39 pKa = 10.17RR40 pKa = 11.84VKK42 pKa = 9.87SRR44 pKa = 11.84SSSRR48 pKa = 11.84VKK50 pKa = 10.63ASRR53 pKa = 11.84SSSATRR59 pKa = 11.84TRR61 pKa = 11.84TRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84GPSAGSAGTDD75 pKa = 3.26GSVSSFRR82 pKa = 11.84YY83 pKa = 8.54SVRR86 pKa = 11.84RR87 pKa = 11.84GVKK90 pKa = 8.91VPKK93 pKa = 10.41GFRR96 pKa = 11.84KK97 pKa = 10.0LLAPRR102 pKa = 11.84HH103 pKa = 5.22MNSVAFSKK111 pKa = 9.27ITSAVNRR118 pKa = 11.84QGVNMVNLFNNGLAPANGTVVEE140 pKa = 4.62AVPSFLDD147 pKa = 3.56TEE149 pKa = 4.82LMMTNLLSDD158 pKa = 4.13EE159 pKa = 4.63AATSVGVNTIRR170 pKa = 11.84YY171 pKa = 6.73WIPFISLKK179 pKa = 8.87ITFRR183 pKa = 11.84SMTSSPVHH191 pKa = 5.67LKK193 pKa = 10.13IYY195 pKa = 9.91NCVARR200 pKa = 11.84RR201 pKa = 11.84DD202 pKa = 3.98QEE204 pKa = 3.9WRR206 pKa = 11.84PTVAFNMANSPYY218 pKa = 7.85TTWSAGLTDD227 pKa = 3.76EE228 pKa = 5.36FGPGSTPTLTVPGTTPFQSEE248 pKa = 5.31SFCQWWNVKK257 pKa = 8.69SSKK260 pKa = 10.37FVMLHH265 pKa = 6.31PGAEE269 pKa = 3.93HH270 pKa = 4.89VHH272 pKa = 6.03FVRR275 pKa = 11.84IQLNRR280 pKa = 11.84LHH282 pKa = 6.64NRR284 pKa = 11.84ALTNDD289 pKa = 3.72LAAIARR295 pKa = 11.84EE296 pKa = 4.56SYY298 pKa = 11.08ACMAVVRR305 pKa = 11.84GGVVQDD311 pKa = 5.34AIATGNVGYY320 pKa = 10.67GPATVDD326 pKa = 3.49VMCDD330 pKa = 3.07YY331 pKa = 11.28HH332 pKa = 7.88MEE334 pKa = 4.14AQAVGKK340 pKa = 9.97NRR342 pKa = 11.84TIVSQYY348 pKa = 10.62QYY350 pKa = 11.21DD351 pKa = 3.96GAITGTGQQVAEE363 pKa = 4.52DD364 pKa = 4.5TDD366 pKa = 3.6TGAPVVNLGG375 pKa = 3.57
MM1 pKa = 7.8AKK3 pKa = 10.03RR4 pKa = 11.84SRR6 pKa = 11.84SRR8 pKa = 11.84SVSRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 10.19LPSNWHH22 pKa = 5.35MASNVARR29 pKa = 11.84AAGTAYY35 pKa = 10.42DD36 pKa = 3.32AYY38 pKa = 10.07KK39 pKa = 10.17RR40 pKa = 11.84VKK42 pKa = 9.87SRR44 pKa = 11.84SSSRR48 pKa = 11.84VKK50 pKa = 10.63ASRR53 pKa = 11.84SSSATRR59 pKa = 11.84TRR61 pKa = 11.84TRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84GPSAGSAGTDD75 pKa = 3.26GSVSSFRR82 pKa = 11.84YY83 pKa = 8.54SVRR86 pKa = 11.84RR87 pKa = 11.84GVKK90 pKa = 8.91VPKK93 pKa = 10.41GFRR96 pKa = 11.84KK97 pKa = 10.0LLAPRR102 pKa = 11.84HH103 pKa = 5.22MNSVAFSKK111 pKa = 9.27ITSAVNRR118 pKa = 11.84QGVNMVNLFNNGLAPANGTVVEE140 pKa = 4.62AVPSFLDD147 pKa = 3.56TEE149 pKa = 4.82LMMTNLLSDD158 pKa = 4.13EE159 pKa = 4.63AATSVGVNTIRR170 pKa = 11.84YY171 pKa = 6.73WIPFISLKK179 pKa = 8.87ITFRR183 pKa = 11.84SMTSSPVHH191 pKa = 5.67LKK193 pKa = 10.13IYY195 pKa = 9.91NCVARR200 pKa = 11.84RR201 pKa = 11.84DD202 pKa = 3.98QEE204 pKa = 3.9WRR206 pKa = 11.84PTVAFNMANSPYY218 pKa = 7.85TTWSAGLTDD227 pKa = 3.76EE228 pKa = 5.36FGPGSTPTLTVPGTTPFQSEE248 pKa = 5.31SFCQWWNVKK257 pKa = 8.69SSKK260 pKa = 10.37FVMLHH265 pKa = 6.31PGAEE269 pKa = 3.93HH270 pKa = 4.89VHH272 pKa = 6.03FVRR275 pKa = 11.84IQLNRR280 pKa = 11.84LHH282 pKa = 6.64NRR284 pKa = 11.84ALTNDD289 pKa = 3.72LAAIARR295 pKa = 11.84EE296 pKa = 4.56SYY298 pKa = 11.08ACMAVVRR305 pKa = 11.84GGVVQDD311 pKa = 5.34AIATGNVGYY320 pKa = 10.67GPATVDD326 pKa = 3.49VMCDD330 pKa = 3.07YY331 pKa = 11.28HH332 pKa = 7.88MEE334 pKa = 4.14AQAVGKK340 pKa = 9.97NRR342 pKa = 11.84TIVSQYY348 pKa = 10.62QYY350 pKa = 11.21DD351 pKa = 3.96GAITGTGQQVAEE363 pKa = 4.52DD364 pKa = 4.5TDD366 pKa = 3.6TGAPVVNLGG375 pKa = 3.57
Molecular weight: 40.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
694 |
319 |
375 |
347.0 |
38.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.79 ± 1.026 | 1.297 ± 0.176 |
4.323 ± 0.654 | 3.746 ± 0.825 |
4.179 ± 0.544 | 7.205 ± 0.2 |
3.458 ± 1.012 | 4.611 ± 1.282 |
3.602 ± 0.1 | 6.916 ± 1.413 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.017 ± 0.903 | 5.764 ± 0.125 |
4.467 ± 0.051 | 3.458 ± 0.401 |
7.781 ± 0.778 | 8.069 ± 1.781 |
7.925 ± 0.057 | 7.637 ± 1.703 |
2.017 ± 0.319 | 2.738 ± 0.149 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
