
Caretta caretta papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyozetapapillomavirus; Dyozetapapillomavirus 1
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
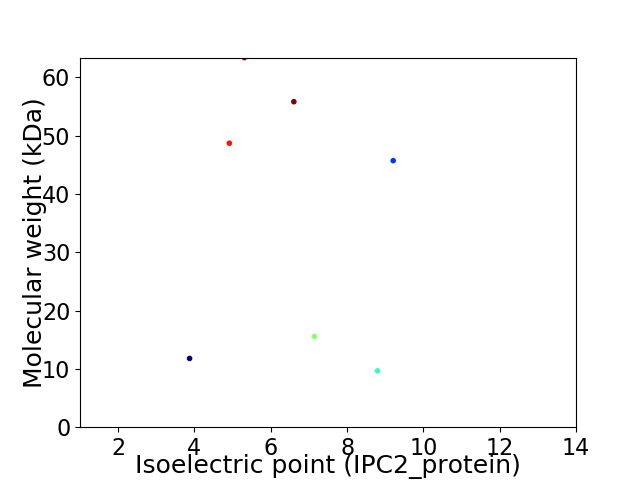
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
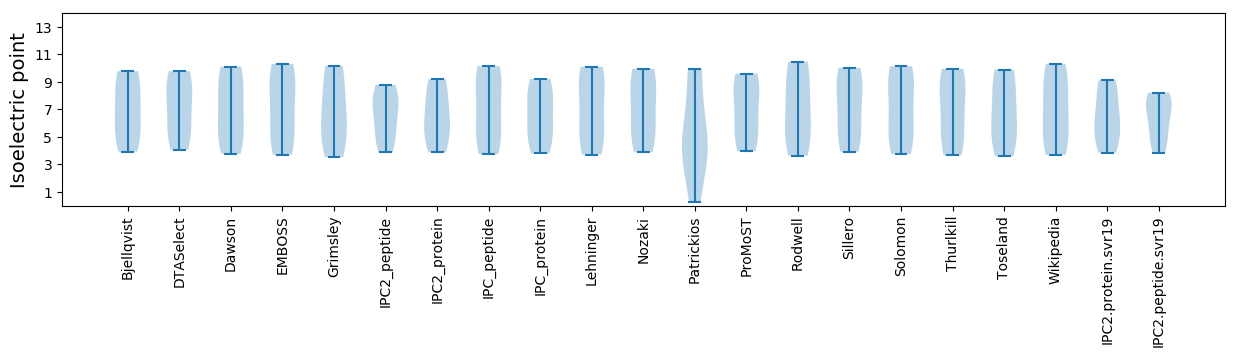
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6RUQ0|B6RUQ0_9PAPI E1 replication protein OS=Caretta caretta papillomavirus 1 OX=485241 GN=E1 PE=4 SV=1
MM1 pKa = 7.42HH2 pKa = 7.45NKK4 pKa = 9.04GQPIKK9 pKa = 10.15GYY11 pKa = 9.24SGKK14 pKa = 10.64YY15 pKa = 8.33MCACCRR21 pKa = 11.84KK22 pKa = 9.73DD23 pKa = 3.42VSFAQQEE30 pKa = 4.12ICIMLSEE37 pKa = 4.06QLGMLVCMNCEE48 pKa = 3.76VVVPSNQIQDD58 pKa = 3.94ALGTADD64 pKa = 3.83LAEE67 pKa = 4.33VTVSCGIGLVDD78 pKa = 4.69EE79 pKa = 5.33GSDD82 pKa = 3.25LASYY86 pKa = 11.01SEE88 pKa = 5.2LEE90 pKa = 4.1TDD92 pKa = 3.04TDD94 pKa = 3.74SDD96 pKa = 4.2NEE98 pKa = 4.29GEE100 pKa = 4.33AFLAGTDD107 pKa = 3.78SEE109 pKa = 4.8AVV111 pKa = 3.23
MM1 pKa = 7.42HH2 pKa = 7.45NKK4 pKa = 9.04GQPIKK9 pKa = 10.15GYY11 pKa = 9.24SGKK14 pKa = 10.64YY15 pKa = 8.33MCACCRR21 pKa = 11.84KK22 pKa = 9.73DD23 pKa = 3.42VSFAQQEE30 pKa = 4.12ICIMLSEE37 pKa = 4.06QLGMLVCMNCEE48 pKa = 3.76VVVPSNQIQDD58 pKa = 3.94ALGTADD64 pKa = 3.83LAEE67 pKa = 4.33VTVSCGIGLVDD78 pKa = 4.69EE79 pKa = 5.33GSDD82 pKa = 3.25LASYY86 pKa = 11.01SEE88 pKa = 5.2LEE90 pKa = 4.1TDD92 pKa = 3.04TDD94 pKa = 3.74SDD96 pKa = 4.2NEE98 pKa = 4.29GEE100 pKa = 4.33AFLAGTDD107 pKa = 3.78SEE109 pKa = 4.8AVV111 pKa = 3.23
Molecular weight: 11.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6RUQ2|B6RUQ2_9PAPI Putative E4 protein OS=Caretta caretta papillomavirus 1 OX=485241 GN=E4 PE=4 SV=1
MM1 pKa = 8.04DD2 pKa = 5.56ANTRR6 pKa = 11.84LMEE9 pKa = 4.28MQDD12 pKa = 3.02KK13 pKa = 10.73QMTIIEE19 pKa = 4.88KK20 pKa = 10.26DD21 pKa = 3.66DD22 pKa = 3.54QSLDD26 pKa = 4.36DD27 pKa = 3.43ILEE30 pKa = 4.12YY31 pKa = 11.13YY32 pKa = 10.42FAVKK36 pKa = 10.43DD37 pKa = 3.86EE38 pKa = 4.0YY39 pKa = 11.33LILAAARR46 pKa = 11.84KK47 pKa = 9.93AGVAHH52 pKa = 7.22IGLQRR57 pKa = 11.84VPSLQVSEE65 pKa = 4.36SKK67 pKa = 10.81YY68 pKa = 10.13RR69 pKa = 11.84EE70 pKa = 4.29AVTMIIIVQSLQNSQFKK87 pKa = 10.33NVNFTLRR94 pKa = 11.84DD95 pKa = 3.47LQYY98 pKa = 10.69QLVMQAPAFTIKK110 pKa = 10.36KK111 pKa = 9.3GPKK114 pKa = 7.78TVYY117 pKa = 8.84VTYY120 pKa = 10.58SGPDD124 pKa = 3.28KK125 pKa = 10.8LTVTHH130 pKa = 6.53TKK132 pKa = 9.5WKK134 pKa = 10.21DD135 pKa = 2.66IYY137 pKa = 9.5YY138 pKa = 10.26QRR140 pKa = 11.84DD141 pKa = 3.65EE142 pKa = 3.91QWRR145 pKa = 11.84SSKK148 pKa = 10.61DD149 pKa = 3.34LQHH152 pKa = 6.1THH154 pKa = 6.85DD155 pKa = 4.81PNWFRR160 pKa = 11.84AHH162 pKa = 6.56TLTDD166 pKa = 3.7SKK168 pKa = 11.63GLFYY172 pKa = 11.11VDD174 pKa = 3.51IYY176 pKa = 11.41GDD178 pKa = 2.95VDD180 pKa = 4.16YY181 pKa = 11.46YY182 pKa = 11.86VLFNSGEE189 pKa = 4.08PQNATRR195 pKa = 11.84KK196 pKa = 8.28GQWEE200 pKa = 4.44ISTSPPDD207 pKa = 3.45ARR209 pKa = 11.84TSTDD213 pKa = 3.0TPSRR217 pKa = 11.84KK218 pKa = 7.58TPEE221 pKa = 3.63RR222 pKa = 11.84HH223 pKa = 5.37IVAITPPIRR232 pKa = 11.84NPRR235 pKa = 11.84NPAYY239 pKa = 9.83QSKK242 pKa = 8.26PTHH245 pKa = 5.78PTPTKK250 pKa = 8.27PTHH253 pKa = 5.98TSTVTTRR260 pKa = 11.84RR261 pKa = 11.84GNGRR265 pKa = 11.84GRR267 pKa = 11.84GLGGRR272 pKa = 11.84GGGGGGAGGSTTRR285 pKa = 11.84RR286 pKa = 11.84PFAEE290 pKa = 4.03KK291 pKa = 10.0QSPVSAEE298 pKa = 3.79EE299 pKa = 3.79VGRR302 pKa = 11.84TRR304 pKa = 11.84EE305 pKa = 4.23TVQGTGTRR313 pKa = 11.84LEE315 pKa = 4.22RR316 pKa = 11.84LLKK319 pKa = 9.98EE320 pKa = 4.38ARR322 pKa = 11.84DD323 pKa = 3.68PPGLVFEE330 pKa = 5.3GSTAQIKK337 pKa = 9.65HH338 pKa = 4.21IRR340 pKa = 11.84RR341 pKa = 11.84RR342 pKa = 11.84VEE344 pKa = 3.94AGSLKK349 pKa = 9.78YY350 pKa = 10.82SRR352 pKa = 11.84VTSTWHH358 pKa = 6.04WISGKK363 pKa = 10.21KK364 pKa = 8.52VLKK367 pKa = 7.84PSKK370 pKa = 10.13MIVVFNNEE378 pKa = 3.96KK379 pKa = 10.28EE380 pKa = 3.98RR381 pKa = 11.84STFLNLFRR389 pKa = 11.84VEE391 pKa = 4.0GDD393 pKa = 3.59GITVRR398 pKa = 11.84LCSFNGLL405 pKa = 3.61
MM1 pKa = 8.04DD2 pKa = 5.56ANTRR6 pKa = 11.84LMEE9 pKa = 4.28MQDD12 pKa = 3.02KK13 pKa = 10.73QMTIIEE19 pKa = 4.88KK20 pKa = 10.26DD21 pKa = 3.66DD22 pKa = 3.54QSLDD26 pKa = 4.36DD27 pKa = 3.43ILEE30 pKa = 4.12YY31 pKa = 11.13YY32 pKa = 10.42FAVKK36 pKa = 10.43DD37 pKa = 3.86EE38 pKa = 4.0YY39 pKa = 11.33LILAAARR46 pKa = 11.84KK47 pKa = 9.93AGVAHH52 pKa = 7.22IGLQRR57 pKa = 11.84VPSLQVSEE65 pKa = 4.36SKK67 pKa = 10.81YY68 pKa = 10.13RR69 pKa = 11.84EE70 pKa = 4.29AVTMIIIVQSLQNSQFKK87 pKa = 10.33NVNFTLRR94 pKa = 11.84DD95 pKa = 3.47LQYY98 pKa = 10.69QLVMQAPAFTIKK110 pKa = 10.36KK111 pKa = 9.3GPKK114 pKa = 7.78TVYY117 pKa = 8.84VTYY120 pKa = 10.58SGPDD124 pKa = 3.28KK125 pKa = 10.8LTVTHH130 pKa = 6.53TKK132 pKa = 9.5WKK134 pKa = 10.21DD135 pKa = 2.66IYY137 pKa = 9.5YY138 pKa = 10.26QRR140 pKa = 11.84DD141 pKa = 3.65EE142 pKa = 3.91QWRR145 pKa = 11.84SSKK148 pKa = 10.61DD149 pKa = 3.34LQHH152 pKa = 6.1THH154 pKa = 6.85DD155 pKa = 4.81PNWFRR160 pKa = 11.84AHH162 pKa = 6.56TLTDD166 pKa = 3.7SKK168 pKa = 11.63GLFYY172 pKa = 11.11VDD174 pKa = 3.51IYY176 pKa = 11.41GDD178 pKa = 2.95VDD180 pKa = 4.16YY181 pKa = 11.46YY182 pKa = 11.86VLFNSGEE189 pKa = 4.08PQNATRR195 pKa = 11.84KK196 pKa = 8.28GQWEE200 pKa = 4.44ISTSPPDD207 pKa = 3.45ARR209 pKa = 11.84TSTDD213 pKa = 3.0TPSRR217 pKa = 11.84KK218 pKa = 7.58TPEE221 pKa = 3.63RR222 pKa = 11.84HH223 pKa = 5.37IVAITPPIRR232 pKa = 11.84NPRR235 pKa = 11.84NPAYY239 pKa = 9.83QSKK242 pKa = 8.26PTHH245 pKa = 5.78PTPTKK250 pKa = 8.27PTHH253 pKa = 5.98TSTVTTRR260 pKa = 11.84RR261 pKa = 11.84GNGRR265 pKa = 11.84GRR267 pKa = 11.84GLGGRR272 pKa = 11.84GGGGGGAGGSTTRR285 pKa = 11.84RR286 pKa = 11.84PFAEE290 pKa = 4.03KK291 pKa = 10.0QSPVSAEE298 pKa = 3.79EE299 pKa = 3.79VGRR302 pKa = 11.84TRR304 pKa = 11.84EE305 pKa = 4.23TVQGTGTRR313 pKa = 11.84LEE315 pKa = 4.22RR316 pKa = 11.84LLKK319 pKa = 9.98EE320 pKa = 4.38ARR322 pKa = 11.84DD323 pKa = 3.68PPGLVFEE330 pKa = 5.3GSTAQIKK337 pKa = 9.65HH338 pKa = 4.21IRR340 pKa = 11.84RR341 pKa = 11.84RR342 pKa = 11.84VEE344 pKa = 3.94AGSLKK349 pKa = 9.78YY350 pKa = 10.82SRR352 pKa = 11.84VTSTWHH358 pKa = 6.04WISGKK363 pKa = 10.21KK364 pKa = 8.52VLKK367 pKa = 7.84PSKK370 pKa = 10.13MIVVFNNEE378 pKa = 3.96KK379 pKa = 10.28EE380 pKa = 3.98RR381 pKa = 11.84STFLNLFRR389 pKa = 11.84VEE391 pKa = 4.0GDD393 pKa = 3.59GITVRR398 pKa = 11.84LCSFNGLL405 pKa = 3.61
Molecular weight: 45.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2254 |
85 |
561 |
322.0 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.61 ± 0.597 | 2.263 ± 0.713 |
6.034 ± 0.44 | 5.457 ± 0.459 |
4.525 ± 0.526 | 6.744 ± 0.664 |
2.085 ± 0.252 | 4.392 ± 0.414 |
5.324 ± 0.681 | 7.941 ± 0.682 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.307 ± 0.255 | 3.86 ± 0.477 |
7.72 ± 1.286 | 3.904 ± 0.488 |
5.723 ± 0.574 | 7.72 ± 0.637 |
6.832 ± 0.749 | 6.3 ± 0.709 |
1.287 ± 0.244 | 2.972 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
