
Wuhan Fly Virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Domestica sigmavirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
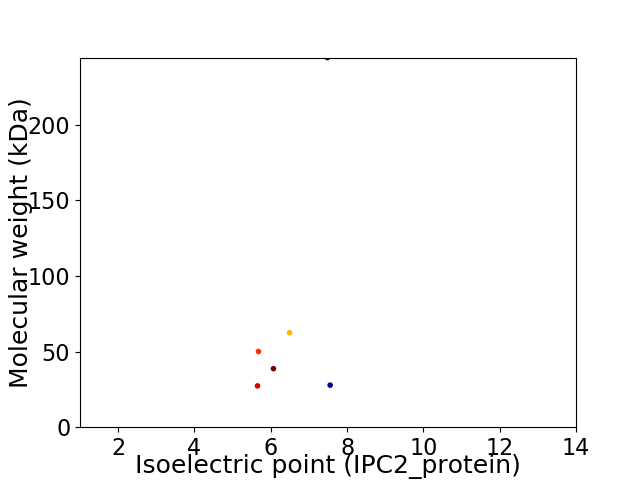
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
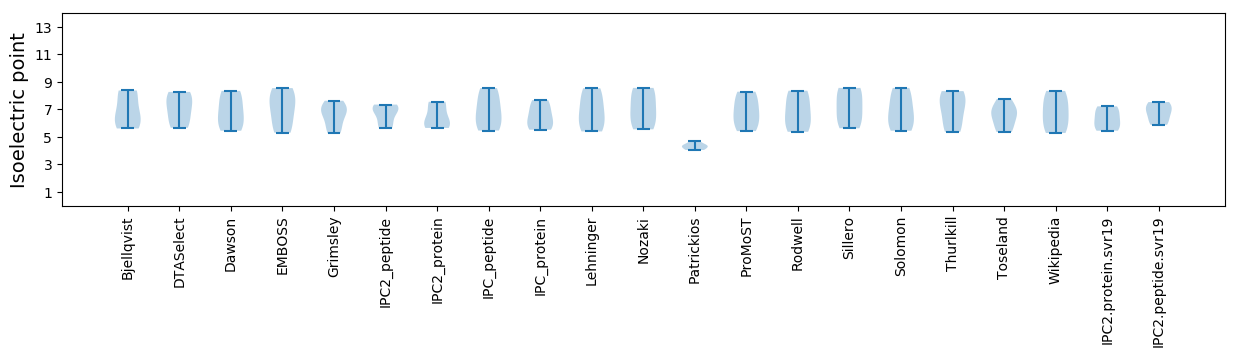
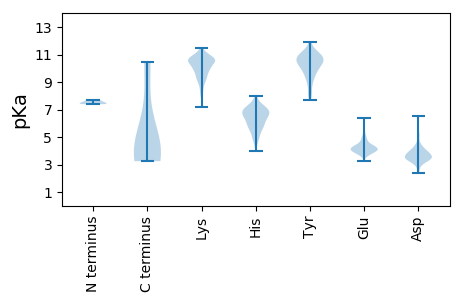
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXI3|A0A0B5KXI3_9RHAB X protein OS=Wuhan Fly Virus 2 OX=1608102 GN=X PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.55KK3 pKa = 9.18MEE5 pKa = 4.21TPRR8 pKa = 11.84GKK10 pKa = 9.96IRR12 pKa = 11.84GRR14 pKa = 11.84VNKK17 pKa = 9.2TRR19 pKa = 11.84VNNKK23 pKa = 9.15DD24 pKa = 3.24VCDD27 pKa = 3.69MLNNRR32 pKa = 11.84GGNLIKK38 pKa = 9.93TVEE41 pKa = 4.06NTLMRR46 pKa = 11.84DD47 pKa = 3.18NDD49 pKa = 3.68MMDD52 pKa = 3.25EE53 pKa = 4.25SEE55 pKa = 4.68APSTSGMNPNSVSDD69 pKa = 4.35PNDD72 pKa = 2.95IPLDD76 pKa = 3.51VLEE79 pKa = 4.62TGKK82 pKa = 10.59EE83 pKa = 3.83LLEE86 pKa = 4.3VVDD89 pKa = 4.13QEE91 pKa = 4.43EE92 pKa = 4.24DD93 pKa = 3.51SEE95 pKa = 4.6MEE97 pKa = 4.13CEE99 pKa = 4.36EE100 pKa = 4.16ILSVSEE106 pKa = 4.13VGGKK110 pKa = 9.97VYY112 pKa = 10.47FSVPPSKK119 pKa = 10.59VWTSEE124 pKa = 3.82QVVMFAAQFMEE135 pKa = 4.91IIKK138 pKa = 10.39GMKK141 pKa = 8.54SEE143 pKa = 5.42KK144 pKa = 8.9MDD146 pKa = 3.96CVPTPSAKK154 pKa = 9.75KK155 pKa = 10.26DD156 pKa = 3.5SPEE159 pKa = 3.99SKK161 pKa = 9.88KK162 pKa = 10.67PKK164 pKa = 8.62ITTPVPSTKK173 pKa = 9.98CDD175 pKa = 2.96ILKK178 pKa = 10.24RR179 pKa = 11.84LSEE182 pKa = 4.21GVVLKK187 pKa = 9.92PIKK190 pKa = 10.14IGLKK194 pKa = 9.47PIRR197 pKa = 11.84VTLDD201 pKa = 3.23NLGVSYY207 pKa = 11.09QEE209 pKa = 4.17AEE211 pKa = 4.44EE212 pKa = 3.91ILKK215 pKa = 10.5RR216 pKa = 11.84LEE218 pKa = 4.18DD219 pKa = 3.61ASIISLIRR227 pKa = 11.84SIKK230 pKa = 9.53ISKK233 pKa = 8.31GQNGRR238 pKa = 11.84FLSNYY243 pKa = 9.66KK244 pKa = 9.89EE245 pKa = 3.98
MM1 pKa = 7.61KK2 pKa = 10.55KK3 pKa = 9.18MEE5 pKa = 4.21TPRR8 pKa = 11.84GKK10 pKa = 9.96IRR12 pKa = 11.84GRR14 pKa = 11.84VNKK17 pKa = 9.2TRR19 pKa = 11.84VNNKK23 pKa = 9.15DD24 pKa = 3.24VCDD27 pKa = 3.69MLNNRR32 pKa = 11.84GGNLIKK38 pKa = 9.93TVEE41 pKa = 4.06NTLMRR46 pKa = 11.84DD47 pKa = 3.18NDD49 pKa = 3.68MMDD52 pKa = 3.25EE53 pKa = 4.25SEE55 pKa = 4.68APSTSGMNPNSVSDD69 pKa = 4.35PNDD72 pKa = 2.95IPLDD76 pKa = 3.51VLEE79 pKa = 4.62TGKK82 pKa = 10.59EE83 pKa = 3.83LLEE86 pKa = 4.3VVDD89 pKa = 4.13QEE91 pKa = 4.43EE92 pKa = 4.24DD93 pKa = 3.51SEE95 pKa = 4.6MEE97 pKa = 4.13CEE99 pKa = 4.36EE100 pKa = 4.16ILSVSEE106 pKa = 4.13VGGKK110 pKa = 9.97VYY112 pKa = 10.47FSVPPSKK119 pKa = 10.59VWTSEE124 pKa = 3.82QVVMFAAQFMEE135 pKa = 4.91IIKK138 pKa = 10.39GMKK141 pKa = 8.54SEE143 pKa = 5.42KK144 pKa = 8.9MDD146 pKa = 3.96CVPTPSAKK154 pKa = 9.75KK155 pKa = 10.26DD156 pKa = 3.5SPEE159 pKa = 3.99SKK161 pKa = 9.88KK162 pKa = 10.67PKK164 pKa = 8.62ITTPVPSTKK173 pKa = 9.98CDD175 pKa = 2.96ILKK178 pKa = 10.24RR179 pKa = 11.84LSEE182 pKa = 4.21GVVLKK187 pKa = 9.92PIKK190 pKa = 10.14IGLKK194 pKa = 9.47PIRR197 pKa = 11.84VTLDD201 pKa = 3.23NLGVSYY207 pKa = 11.09QEE209 pKa = 4.17AEE211 pKa = 4.44EE212 pKa = 3.91ILKK215 pKa = 10.5RR216 pKa = 11.84LEE218 pKa = 4.18DD219 pKa = 3.61ASIISLIRR227 pKa = 11.84SIKK230 pKa = 9.53ISKK233 pKa = 8.31GQNGRR238 pKa = 11.84FLSNYY243 pKa = 9.66KK244 pKa = 9.89EE245 pKa = 3.98
Molecular weight: 27.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KK73|A0A0B5KK73_9RHAB Glycoprotein OS=Wuhan Fly Virus 2 OX=1608102 GN=G PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 9.16KK3 pKa = 8.88TNTEE7 pKa = 3.41IKK9 pKa = 9.56TPPTSKK15 pKa = 10.34MNFNNFLSSFGKK27 pKa = 10.33QKK29 pKa = 10.77GSPEE33 pKa = 3.93KK34 pKa = 10.23RR35 pKa = 11.84DD36 pKa = 3.61LSVGLYY42 pKa = 10.27GPSAPFEE49 pKa = 4.67SISDD53 pKa = 3.7SLNVFEE59 pKa = 4.73YY60 pKa = 10.82TEE62 pKa = 4.37DD63 pKa = 3.59DD64 pKa = 3.96YY65 pKa = 11.94RR66 pKa = 11.84PLPEE70 pKa = 4.96TIPDD74 pKa = 3.56VSKK77 pKa = 8.23TWSVVGRR84 pKa = 11.84MEE86 pKa = 4.02YY87 pKa = 9.23LTNRR91 pKa = 11.84KK92 pKa = 8.88VSSWEE97 pKa = 3.96DD98 pKa = 3.32VLEE101 pKa = 4.56DD102 pKa = 4.26LDD104 pKa = 5.12VILDD108 pKa = 3.98EE109 pKa = 4.42YY110 pKa = 10.64RR111 pKa = 11.84GSVLYY116 pKa = 10.6RR117 pKa = 11.84EE118 pKa = 4.39IFIIFYY124 pKa = 10.49YY125 pKa = 11.11LMGTHH130 pKa = 6.98LRR132 pKa = 11.84PVQSPGGGLRR142 pKa = 11.84KK143 pKa = 9.52FRR145 pKa = 11.84IDD147 pKa = 2.99MEE149 pKa = 4.18ARR151 pKa = 11.84VNLVHH156 pKa = 7.31GFKK159 pKa = 10.63LDD161 pKa = 3.52NDD163 pKa = 3.94KK164 pKa = 11.11EE165 pKa = 4.25KK166 pKa = 10.93QINWTYY172 pKa = 11.92DD173 pKa = 2.94SMKK176 pKa = 10.31SIRR179 pKa = 11.84KK180 pKa = 8.81CRR182 pKa = 11.84ITYY185 pKa = 7.69MCSLKK190 pKa = 9.98DD191 pKa = 3.65TNRR194 pKa = 11.84KK195 pKa = 8.83GMPFKK200 pKa = 10.64TMYY203 pKa = 10.36LYY205 pKa = 10.23PSLEE209 pKa = 4.16GTPTPDD215 pKa = 4.15LKK217 pKa = 11.23KK218 pKa = 10.86LLDD221 pKa = 3.34PHH223 pKa = 7.01NIFFEE228 pKa = 4.49DD229 pKa = 3.35LGDD232 pKa = 3.41NYY234 pKa = 10.83RR235 pKa = 11.84FYY237 pKa = 11.31KK238 pKa = 10.44
MM1 pKa = 7.69KK2 pKa = 9.16KK3 pKa = 8.88TNTEE7 pKa = 3.41IKK9 pKa = 9.56TPPTSKK15 pKa = 10.34MNFNNFLSSFGKK27 pKa = 10.33QKK29 pKa = 10.77GSPEE33 pKa = 3.93KK34 pKa = 10.23RR35 pKa = 11.84DD36 pKa = 3.61LSVGLYY42 pKa = 10.27GPSAPFEE49 pKa = 4.67SISDD53 pKa = 3.7SLNVFEE59 pKa = 4.73YY60 pKa = 10.82TEE62 pKa = 4.37DD63 pKa = 3.59DD64 pKa = 3.96YY65 pKa = 11.94RR66 pKa = 11.84PLPEE70 pKa = 4.96TIPDD74 pKa = 3.56VSKK77 pKa = 8.23TWSVVGRR84 pKa = 11.84MEE86 pKa = 4.02YY87 pKa = 9.23LTNRR91 pKa = 11.84KK92 pKa = 8.88VSSWEE97 pKa = 3.96DD98 pKa = 3.32VLEE101 pKa = 4.56DD102 pKa = 4.26LDD104 pKa = 5.12VILDD108 pKa = 3.98EE109 pKa = 4.42YY110 pKa = 10.64RR111 pKa = 11.84GSVLYY116 pKa = 10.6RR117 pKa = 11.84EE118 pKa = 4.39IFIIFYY124 pKa = 10.49YY125 pKa = 11.11LMGTHH130 pKa = 6.98LRR132 pKa = 11.84PVQSPGGGLRR142 pKa = 11.84KK143 pKa = 9.52FRR145 pKa = 11.84IDD147 pKa = 2.99MEE149 pKa = 4.18ARR151 pKa = 11.84VNLVHH156 pKa = 7.31GFKK159 pKa = 10.63LDD161 pKa = 3.52NDD163 pKa = 3.94KK164 pKa = 11.11EE165 pKa = 4.25KK166 pKa = 10.93QINWTYY172 pKa = 11.92DD173 pKa = 2.94SMKK176 pKa = 10.31SIRR179 pKa = 11.84KK180 pKa = 8.81CRR182 pKa = 11.84ITYY185 pKa = 7.69MCSLKK190 pKa = 9.98DD191 pKa = 3.65TNRR194 pKa = 11.84KK195 pKa = 8.83GMPFKK200 pKa = 10.64TMYY203 pKa = 10.36LYY205 pKa = 10.23PSLEE209 pKa = 4.16GTPTPDD215 pKa = 4.15LKK217 pKa = 11.23KK218 pKa = 10.86LLDD221 pKa = 3.34PHH223 pKa = 7.01NIFFEE228 pKa = 4.49DD229 pKa = 3.35LGDD232 pKa = 3.41NYY234 pKa = 10.83RR235 pKa = 11.84FYY237 pKa = 11.31KK238 pKa = 10.44
Molecular weight: 27.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3938 |
238 |
2134 |
656.3 |
75.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.723 ± 0.427 | 1.549 ± 0.18 |
5.637 ± 0.279 | 6.374 ± 0.509 |
4.545 ± 0.272 | 5.053 ± 0.46 |
2.539 ± 0.442 | 6.475 ± 0.426 |
6.882 ± 0.412 | 9.751 ± 1.172 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.996 ± 0.381 | 5.079 ± 0.186 |
4.647 ± 0.398 | 2.996 ± 0.243 |
5.256 ± 0.142 | 7.923 ± 0.306 |
6.069 ± 0.338 | 6.094 ± 0.457 |
1.549 ± 0.246 | 3.86 ± 0.338 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
