
Roseivivax roseus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseivivax
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
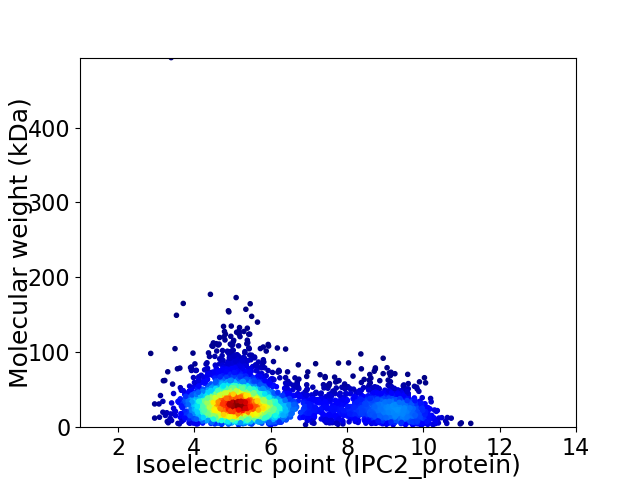
Virtual 2D-PAGE plot for 4061 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9P807|A0A1H9P807_9RHOB Trk system potassium uptake protein TrkA OS=Roseivivax roseus OX=641238 GN=SAMN04490244_10159 PE=4 SV=1
MM1 pKa = 7.3KK2 pKa = 10.44RR3 pKa = 11.84IATLTALGALIAGQAGAGGVEE24 pKa = 4.85RR25 pKa = 11.84SAQSAGILFEE35 pKa = 6.52DD36 pKa = 4.54GDD38 pKa = 4.14RR39 pKa = 11.84VEE41 pKa = 5.51LSFGYY46 pKa = 10.31VSPDD50 pKa = 3.06VSGEE54 pKa = 3.7QLLPVGPTSPAGASSGDD71 pKa = 3.37MSEE74 pKa = 5.42DD75 pKa = 3.57YY76 pKa = 10.11STLAFGYY83 pKa = 8.7KK84 pKa = 9.01QQVNEE89 pKa = 4.35SVALALIVDD98 pKa = 4.12NPIGADD104 pKa = 3.34VNYY107 pKa = 10.77GSDD110 pKa = 3.05TQYY113 pKa = 11.36LYY115 pKa = 11.45GGGAPFAALGLNSSTAEE132 pKa = 3.76IDD134 pKa = 3.74SAAVTALASYY144 pKa = 10.12KK145 pKa = 10.56LPSNVTLFGGLRR157 pKa = 11.84AQQASGGVSLFNGYY171 pKa = 8.36EE172 pKa = 4.03MEE174 pKa = 4.45TSSEE178 pKa = 3.65TDD180 pKa = 2.96YY181 pKa = 11.62GYY183 pKa = 11.61VLGAAYY189 pKa = 7.81EE190 pKa = 4.2RR191 pKa = 11.84PDD193 pKa = 2.95IALRR197 pKa = 11.84VALTYY202 pKa = 10.71NSEE205 pKa = 3.88ITHH208 pKa = 6.77DD209 pKa = 4.72FSASEE214 pKa = 4.36TVFDD218 pKa = 4.85PATQSVSNFDD228 pKa = 3.47TSFEE232 pKa = 4.41TTVPQSLNLEE242 pKa = 4.39VQSGIAPDD250 pKa = 3.66TLAFGSVRR258 pKa = 11.84WVDD261 pKa = 2.97WSEE264 pKa = 3.69FDD266 pKa = 3.7ITPSVYY272 pKa = 8.49NTAYY276 pKa = 10.62NGSLVNYY283 pKa = 9.46DD284 pKa = 4.58DD285 pKa = 6.02DD286 pKa = 5.25VITYY290 pKa = 7.77TVGLGRR296 pKa = 11.84RR297 pKa = 11.84FTEE300 pKa = 3.78NWSGAVLATYY310 pKa = 10.06EE311 pKa = 4.12DD312 pKa = 4.07QNGGFSGNLGPTDD325 pKa = 3.68GQTSVGVAVTYY336 pKa = 7.39EE337 pKa = 3.99TEE339 pKa = 4.07RR340 pKa = 11.84YY341 pKa = 8.76EE342 pKa = 3.73ITGGLRR348 pKa = 11.84HH349 pKa = 6.04IWIGDD354 pKa = 3.24ARR356 pKa = 11.84TEE358 pKa = 4.12APSALDD364 pKa = 3.52QPAGTAFGDD373 pKa = 3.86FDD375 pKa = 6.37DD376 pKa = 4.34NTAIAGGLQVAIKK389 pKa = 9.78FF390 pKa = 3.92
MM1 pKa = 7.3KK2 pKa = 10.44RR3 pKa = 11.84IATLTALGALIAGQAGAGGVEE24 pKa = 4.85RR25 pKa = 11.84SAQSAGILFEE35 pKa = 6.52DD36 pKa = 4.54GDD38 pKa = 4.14RR39 pKa = 11.84VEE41 pKa = 5.51LSFGYY46 pKa = 10.31VSPDD50 pKa = 3.06VSGEE54 pKa = 3.7QLLPVGPTSPAGASSGDD71 pKa = 3.37MSEE74 pKa = 5.42DD75 pKa = 3.57YY76 pKa = 10.11STLAFGYY83 pKa = 8.7KK84 pKa = 9.01QQVNEE89 pKa = 4.35SVALALIVDD98 pKa = 4.12NPIGADD104 pKa = 3.34VNYY107 pKa = 10.77GSDD110 pKa = 3.05TQYY113 pKa = 11.36LYY115 pKa = 11.45GGGAPFAALGLNSSTAEE132 pKa = 3.76IDD134 pKa = 3.74SAAVTALASYY144 pKa = 10.12KK145 pKa = 10.56LPSNVTLFGGLRR157 pKa = 11.84AQQASGGVSLFNGYY171 pKa = 8.36EE172 pKa = 4.03MEE174 pKa = 4.45TSSEE178 pKa = 3.65TDD180 pKa = 2.96YY181 pKa = 11.62GYY183 pKa = 11.61VLGAAYY189 pKa = 7.81EE190 pKa = 4.2RR191 pKa = 11.84PDD193 pKa = 2.95IALRR197 pKa = 11.84VALTYY202 pKa = 10.71NSEE205 pKa = 3.88ITHH208 pKa = 6.77DD209 pKa = 4.72FSASEE214 pKa = 4.36TVFDD218 pKa = 4.85PATQSVSNFDD228 pKa = 3.47TSFEE232 pKa = 4.41TTVPQSLNLEE242 pKa = 4.39VQSGIAPDD250 pKa = 3.66TLAFGSVRR258 pKa = 11.84WVDD261 pKa = 2.97WSEE264 pKa = 3.69FDD266 pKa = 3.7ITPSVYY272 pKa = 8.49NTAYY276 pKa = 10.62NGSLVNYY283 pKa = 9.46DD284 pKa = 4.58DD285 pKa = 6.02DD286 pKa = 5.25VITYY290 pKa = 7.77TVGLGRR296 pKa = 11.84RR297 pKa = 11.84FTEE300 pKa = 3.78NWSGAVLATYY310 pKa = 10.06EE311 pKa = 4.12DD312 pKa = 4.07QNGGFSGNLGPTDD325 pKa = 3.68GQTSVGVAVTYY336 pKa = 7.39EE337 pKa = 3.99TEE339 pKa = 4.07RR340 pKa = 11.84YY341 pKa = 8.76EE342 pKa = 3.73ITGGLRR348 pKa = 11.84HH349 pKa = 6.04IWIGDD354 pKa = 3.24ARR356 pKa = 11.84TEE358 pKa = 4.12APSALDD364 pKa = 3.52QPAGTAFGDD373 pKa = 3.86FDD375 pKa = 6.37DD376 pKa = 4.34NTAIAGGLQVAIKK389 pKa = 9.78FF390 pKa = 3.92
Molecular weight: 40.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9XBE8|A0A1H9XBE8_9RHOB HNH endonuclease OS=Roseivivax roseus OX=641238 GN=SAMN04490244_1311 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3AGRR28 pKa = 11.84KK29 pKa = 8.43IVNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 6.6TVSAA44 pKa = 4.51
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3AGRR28 pKa = 11.84KK29 pKa = 8.43IVNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 6.6TVSAA44 pKa = 4.51
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1273032 |
27 |
4772 |
313.5 |
33.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.716 ± 0.06 | 0.848 ± 0.015 |
6.399 ± 0.039 | 6.201 ± 0.041 |
3.539 ± 0.028 | 9.032 ± 0.045 |
2.0 ± 0.02 | 4.683 ± 0.03 |
2.365 ± 0.034 | 10.08 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.638 ± 0.019 | 2.221 ± 0.021 |
5.26 ± 0.027 | 2.938 ± 0.026 |
7.418 ± 0.053 | 5.076 ± 0.026 |
5.581 ± 0.027 | 7.479 ± 0.03 |
1.414 ± 0.018 | 2.111 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
