
Odonata-associated circular virus-13
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.19
Get precalculated fractions of proteins
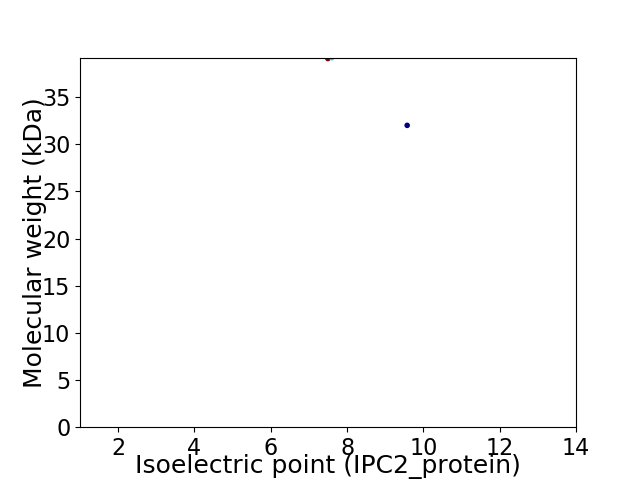
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGT7|A0A0B4UGT7_9VIRU Putative capsid protein OS=Odonata-associated circular virus-13 OX=1592113 PE=4 SV=1
MM1 pKa = 7.48GSNVLRR7 pKa = 11.84AKK9 pKa = 10.37NGSSGPRR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 10.16YY19 pKa = 10.64KK20 pKa = 10.48LYY22 pKa = 10.34CFTSFAVDD30 pKa = 3.25QEE32 pKa = 4.22PSYY35 pKa = 11.61DD36 pKa = 3.73PTAMQYY42 pKa = 10.92LIYY45 pKa = 10.62GRR47 pKa = 11.84EE48 pKa = 3.89TCPDD52 pKa = 3.1SGRR55 pKa = 11.84THH57 pKa = 6.61LQGFVAFKK65 pKa = 9.95NRR67 pKa = 11.84QYY69 pKa = 10.15FTACKK74 pKa = 9.76KK75 pKa = 10.65YY76 pKa = 10.13FGTAHH81 pKa = 6.45VEE83 pKa = 3.95ACKK86 pKa = 9.95GTFAEE91 pKa = 4.12NQEE94 pKa = 4.2YY95 pKa = 10.2CKK97 pKa = 10.6KK98 pKa = 10.86DD99 pKa = 3.0GDD101 pKa = 3.97YY102 pKa = 11.34KK103 pKa = 11.18EE104 pKa = 5.56FGTAPEE110 pKa = 4.32IKK112 pKa = 10.34SGGDD116 pKa = 3.09VFNDD120 pKa = 3.29VLRR123 pKa = 11.84KK124 pKa = 10.15AEE126 pKa = 4.3AGSIQEE132 pKa = 4.76IKK134 pKa = 10.65DD135 pKa = 4.13LYY137 pKa = 8.17PGLYY141 pKa = 9.27IRR143 pKa = 11.84YY144 pKa = 8.93KK145 pKa = 9.67KK146 pKa = 8.93TLEE149 pKa = 4.07SIKK152 pKa = 10.52RR153 pKa = 11.84FNAEE157 pKa = 3.91QLEE160 pKa = 4.45EE161 pKa = 4.28SCGIWLTGPPRR172 pKa = 11.84SGKK175 pKa = 10.15DD176 pKa = 3.33YY177 pKa = 11.02AVSTFFSSIYY187 pKa = 10.94SKK189 pKa = 10.34MLNKK193 pKa = 9.62WFDD196 pKa = 3.44GYY198 pKa = 11.04EE199 pKa = 4.13GEE201 pKa = 4.5EE202 pKa = 4.3CVHH205 pKa = 6.9LSDD208 pKa = 3.54MDD210 pKa = 3.98KK211 pKa = 10.36NHH213 pKa = 6.3VYY215 pKa = 9.72MGSFLKK221 pKa = 10.15IWCDD225 pKa = 3.14RR226 pKa = 11.84YY227 pKa = 10.41PFRR230 pKa = 11.84AEE232 pKa = 3.65IKK234 pKa = 9.86GGTMVIRR241 pKa = 11.84PKK243 pKa = 10.88YY244 pKa = 9.87IVVTSNYY251 pKa = 9.48KK252 pKa = 10.67LEE254 pKa = 5.3DD255 pKa = 3.44IFDD258 pKa = 3.97GSMLSALQARR268 pKa = 11.84FMVMCYY274 pKa = 10.59DD275 pKa = 3.58PVDD278 pKa = 3.97GVVVTPRR285 pKa = 11.84PVFQPSDD292 pKa = 3.33RR293 pKa = 11.84FIQALQHH300 pKa = 6.68AIPKK304 pKa = 10.41GEE306 pKa = 5.01DD307 pKa = 3.42VPDD310 pKa = 3.81MAPKK314 pKa = 10.43AGPSNALQEE323 pKa = 4.78AEE325 pKa = 4.52FSSSDD330 pKa = 3.45EE331 pKa = 4.35FKK333 pKa = 10.75EE334 pKa = 4.16SQVPKK339 pKa = 10.2RR340 pKa = 11.84KK341 pKa = 9.83KK342 pKa = 10.31KK343 pKa = 10.39LL344 pKa = 3.32
MM1 pKa = 7.48GSNVLRR7 pKa = 11.84AKK9 pKa = 10.37NGSSGPRR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 10.16YY19 pKa = 10.64KK20 pKa = 10.48LYY22 pKa = 10.34CFTSFAVDD30 pKa = 3.25QEE32 pKa = 4.22PSYY35 pKa = 11.61DD36 pKa = 3.73PTAMQYY42 pKa = 10.92LIYY45 pKa = 10.62GRR47 pKa = 11.84EE48 pKa = 3.89TCPDD52 pKa = 3.1SGRR55 pKa = 11.84THH57 pKa = 6.61LQGFVAFKK65 pKa = 9.95NRR67 pKa = 11.84QYY69 pKa = 10.15FTACKK74 pKa = 9.76KK75 pKa = 10.65YY76 pKa = 10.13FGTAHH81 pKa = 6.45VEE83 pKa = 3.95ACKK86 pKa = 9.95GTFAEE91 pKa = 4.12NQEE94 pKa = 4.2YY95 pKa = 10.2CKK97 pKa = 10.6KK98 pKa = 10.86DD99 pKa = 3.0GDD101 pKa = 3.97YY102 pKa = 11.34KK103 pKa = 11.18EE104 pKa = 5.56FGTAPEE110 pKa = 4.32IKK112 pKa = 10.34SGGDD116 pKa = 3.09VFNDD120 pKa = 3.29VLRR123 pKa = 11.84KK124 pKa = 10.15AEE126 pKa = 4.3AGSIQEE132 pKa = 4.76IKK134 pKa = 10.65DD135 pKa = 4.13LYY137 pKa = 8.17PGLYY141 pKa = 9.27IRR143 pKa = 11.84YY144 pKa = 8.93KK145 pKa = 9.67KK146 pKa = 8.93TLEE149 pKa = 4.07SIKK152 pKa = 10.52RR153 pKa = 11.84FNAEE157 pKa = 3.91QLEE160 pKa = 4.45EE161 pKa = 4.28SCGIWLTGPPRR172 pKa = 11.84SGKK175 pKa = 10.15DD176 pKa = 3.33YY177 pKa = 11.02AVSTFFSSIYY187 pKa = 10.94SKK189 pKa = 10.34MLNKK193 pKa = 9.62WFDD196 pKa = 3.44GYY198 pKa = 11.04EE199 pKa = 4.13GEE201 pKa = 4.5EE202 pKa = 4.3CVHH205 pKa = 6.9LSDD208 pKa = 3.54MDD210 pKa = 3.98KK211 pKa = 10.36NHH213 pKa = 6.3VYY215 pKa = 9.72MGSFLKK221 pKa = 10.15IWCDD225 pKa = 3.14RR226 pKa = 11.84YY227 pKa = 10.41PFRR230 pKa = 11.84AEE232 pKa = 3.65IKK234 pKa = 9.86GGTMVIRR241 pKa = 11.84PKK243 pKa = 10.88YY244 pKa = 9.87IVVTSNYY251 pKa = 9.48KK252 pKa = 10.67LEE254 pKa = 5.3DD255 pKa = 3.44IFDD258 pKa = 3.97GSMLSALQARR268 pKa = 11.84FMVMCYY274 pKa = 10.59DD275 pKa = 3.58PVDD278 pKa = 3.97GVVVTPRR285 pKa = 11.84PVFQPSDD292 pKa = 3.33RR293 pKa = 11.84FIQALQHH300 pKa = 6.68AIPKK304 pKa = 10.41GEE306 pKa = 5.01DD307 pKa = 3.42VPDD310 pKa = 3.81MAPKK314 pKa = 10.43AGPSNALQEE323 pKa = 4.78AEE325 pKa = 4.52FSSSDD330 pKa = 3.45EE331 pKa = 4.35FKK333 pKa = 10.75EE334 pKa = 4.16SQVPKK339 pKa = 10.2RR340 pKa = 11.84KK341 pKa = 9.83KK342 pKa = 10.31KK343 pKa = 10.39LL344 pKa = 3.32
Molecular weight: 39.08 kDa
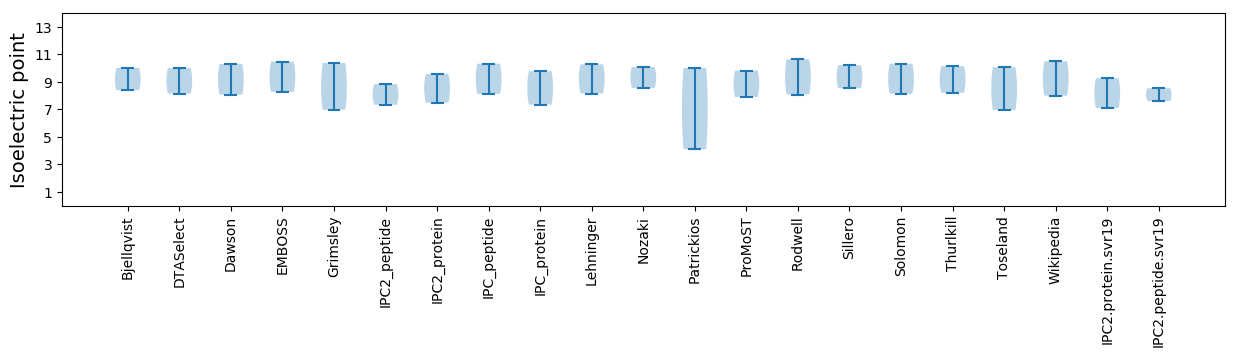
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGT7|A0A0B4UGT7_9VIRU Putative capsid protein OS=Odonata-associated circular virus-13 OX=1592113 PE=4 SV=1
MM1 pKa = 8.19RR2 pKa = 11.84YY3 pKa = 9.5RR4 pKa = 11.84RR5 pKa = 11.84GKK7 pKa = 8.38TYY9 pKa = 9.4RR10 pKa = 11.84TWRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.32RR16 pKa = 11.84GRR18 pKa = 11.84LTRR21 pKa = 11.84YY22 pKa = 8.74KK23 pKa = 10.38KK24 pKa = 10.31RR25 pKa = 11.84NFRR28 pKa = 11.84LRR30 pKa = 11.84MNSRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84FQNEE41 pKa = 3.2RR42 pKa = 11.84RR43 pKa = 11.84SFKK46 pKa = 10.22STATYY51 pKa = 10.67QLFYY55 pKa = 10.66TVDD58 pKa = 4.23LPTSDD63 pKa = 3.7TPTVSNARR71 pKa = 11.84YY72 pKa = 8.55FQDD75 pKa = 3.39SVKK78 pKa = 9.72MFPLGSQLFNRR89 pKa = 11.84YY90 pKa = 8.78LRR92 pKa = 11.84TYY94 pKa = 10.87AWVKK98 pKa = 10.27LNSVTYY104 pKa = 7.45YY105 pKa = 9.8WRR107 pKa = 11.84IGFVGYY113 pKa = 10.82NEE115 pKa = 4.36AEE117 pKa = 4.37KK118 pKa = 11.12VPDD121 pKa = 4.54PNDD124 pKa = 3.69PQKK127 pKa = 11.05KK128 pKa = 7.97KK129 pKa = 10.57QLVVMEE135 pKa = 4.8AVNSLMGKK143 pKa = 9.95FPFYY147 pKa = 11.03LNWSLDD153 pKa = 3.13GDD155 pKa = 4.43NPTKK159 pKa = 10.33MAMDD163 pKa = 5.15DD164 pKa = 3.8LAACPSSRR172 pKa = 11.84KK173 pKa = 10.07VYY175 pKa = 10.68INGKK179 pKa = 9.12KK180 pKa = 8.46ATKK183 pKa = 8.56FTYY186 pKa = 10.18SIPKK190 pKa = 9.07AYY192 pKa = 10.33RR193 pKa = 11.84MYY195 pKa = 10.76LSSSVFRR202 pKa = 11.84SMSVSSGIKK211 pKa = 10.46NNITTLFNNSSLRR224 pKa = 11.84CPDD227 pKa = 3.25KK228 pKa = 11.34FNGSMLDD235 pKa = 3.04IFEE238 pKa = 4.85KK239 pKa = 10.51YY240 pKa = 10.63AGVSLDD246 pKa = 3.58NDD248 pKa = 3.92MACEE252 pKa = 4.63FILWCDD258 pKa = 3.25VYY260 pKa = 10.57MDD262 pKa = 4.56CSFKK266 pKa = 11.1GRR268 pKa = 11.84IDD270 pKa = 3.47NAA272 pKa = 3.81
MM1 pKa = 8.19RR2 pKa = 11.84YY3 pKa = 9.5RR4 pKa = 11.84RR5 pKa = 11.84GKK7 pKa = 8.38TYY9 pKa = 9.4RR10 pKa = 11.84TWRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.32RR16 pKa = 11.84GRR18 pKa = 11.84LTRR21 pKa = 11.84YY22 pKa = 8.74KK23 pKa = 10.38KK24 pKa = 10.31RR25 pKa = 11.84NFRR28 pKa = 11.84LRR30 pKa = 11.84MNSRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84FQNEE41 pKa = 3.2RR42 pKa = 11.84RR43 pKa = 11.84SFKK46 pKa = 10.22STATYY51 pKa = 10.67QLFYY55 pKa = 10.66TVDD58 pKa = 4.23LPTSDD63 pKa = 3.7TPTVSNARR71 pKa = 11.84YY72 pKa = 8.55FQDD75 pKa = 3.39SVKK78 pKa = 9.72MFPLGSQLFNRR89 pKa = 11.84YY90 pKa = 8.78LRR92 pKa = 11.84TYY94 pKa = 10.87AWVKK98 pKa = 10.27LNSVTYY104 pKa = 7.45YY105 pKa = 9.8WRR107 pKa = 11.84IGFVGYY113 pKa = 10.82NEE115 pKa = 4.36AEE117 pKa = 4.37KK118 pKa = 11.12VPDD121 pKa = 4.54PNDD124 pKa = 3.69PQKK127 pKa = 11.05KK128 pKa = 7.97KK129 pKa = 10.57QLVVMEE135 pKa = 4.8AVNSLMGKK143 pKa = 9.95FPFYY147 pKa = 11.03LNWSLDD153 pKa = 3.13GDD155 pKa = 4.43NPTKK159 pKa = 10.33MAMDD163 pKa = 5.15DD164 pKa = 3.8LAACPSSRR172 pKa = 11.84KK173 pKa = 10.07VYY175 pKa = 10.68INGKK179 pKa = 9.12KK180 pKa = 8.46ATKK183 pKa = 8.56FTYY186 pKa = 10.18SIPKK190 pKa = 9.07AYY192 pKa = 10.33RR193 pKa = 11.84MYY195 pKa = 10.76LSSSVFRR202 pKa = 11.84SMSVSSGIKK211 pKa = 10.46NNITTLFNNSSLRR224 pKa = 11.84CPDD227 pKa = 3.25KK228 pKa = 11.34FNGSMLDD235 pKa = 3.04IFEE238 pKa = 4.85KK239 pKa = 10.51YY240 pKa = 10.63AGVSLDD246 pKa = 3.58NDD248 pKa = 3.92MACEE252 pKa = 4.63FILWCDD258 pKa = 3.25VYY260 pKa = 10.57MDD262 pKa = 4.56CSFKK266 pKa = 11.1GRR268 pKa = 11.84IDD270 pKa = 3.47NAA272 pKa = 3.81
Molecular weight: 32.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
616 |
272 |
344 |
308.0 |
35.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.844 ± 0.443 | 2.273 ± 0.276 |
6.006 ± 0.079 | 4.708 ± 1.589 |
6.169 ± 0.052 | 6.169 ± 1.116 |
0.812 ± 0.516 | 3.734 ± 0.504 |
8.604 ± 0.328 | 6.169 ± 0.519 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.571 ± 0.534 | 4.708 ± 1.447 |
4.87 ± 0.525 | 3.084 ± 0.558 |
6.981 ± 1.638 | 8.279 ± 0.579 |
4.87 ± 0.643 | 5.682 ± 0.106 |
1.299 ± 0.343 | 6.169 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
