
Rousettus aegyptiacus adenovirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus; unclassified Mastadenovirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
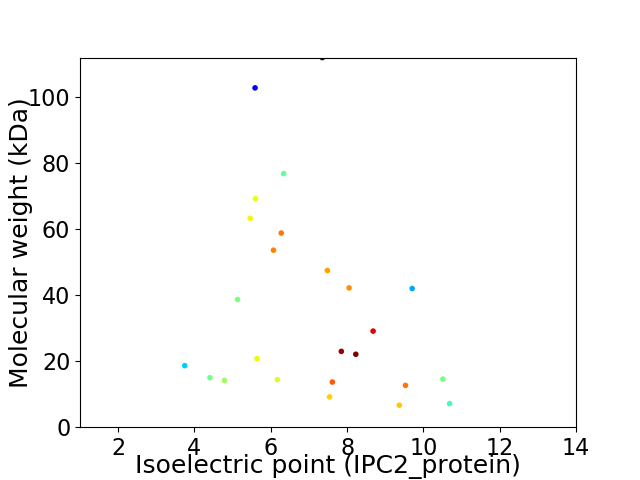
Virtual 2D-PAGE plot for 25 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A344X9T9|A0A344X9T9_9ADEN E1B protein small T-antigen OS=Rousettus aegyptiacus adenovirus OX=1967296 PE=3 SV=1
MM1 pKa = 7.61RR2 pKa = 11.84HH3 pKa = 6.07FSLTCLDD10 pKa = 4.92PIYY13 pKa = 11.0SVADD17 pKa = 3.61DD18 pKa = 4.67FLASLHH24 pKa = 6.32SFSEE28 pKa = 4.44EE29 pKa = 3.87PDD31 pKa = 3.13LSFYY35 pKa = 11.0SDD37 pKa = 3.04SAAPSTSHH45 pKa = 6.74SLPDD49 pKa = 5.03LVDD52 pKa = 4.48LDD54 pKa = 4.3LAPFVPLDD62 pKa = 3.35NYY64 pKa = 10.85VDD66 pKa = 4.61FGYY69 pKa = 10.96EE70 pKa = 3.74SDD72 pKa = 4.3EE73 pKa = 4.5DD74 pKa = 4.25FGLQTVPNLPPSVLLFCDD92 pKa = 4.27APASFPDD99 pKa = 3.33RR100 pKa = 11.84TEE102 pKa = 4.15IDD104 pKa = 3.45LHH106 pKa = 8.45CYY108 pKa = 10.27EE109 pKa = 5.02SMPSSVEE116 pKa = 3.79STGAISPPPSAQVSSSPSDD135 pKa = 3.8CDD137 pKa = 4.02QHH139 pKa = 8.18ICDD142 pKa = 5.26PGNSPCSLCFLRR154 pKa = 11.84LAYY157 pKa = 10.57SGMCFSYY164 pKa = 10.2TFSHH168 pKa = 7.15LLCC171 pKa = 5.17
MM1 pKa = 7.61RR2 pKa = 11.84HH3 pKa = 6.07FSLTCLDD10 pKa = 4.92PIYY13 pKa = 11.0SVADD17 pKa = 3.61DD18 pKa = 4.67FLASLHH24 pKa = 6.32SFSEE28 pKa = 4.44EE29 pKa = 3.87PDD31 pKa = 3.13LSFYY35 pKa = 11.0SDD37 pKa = 3.04SAAPSTSHH45 pKa = 6.74SLPDD49 pKa = 5.03LVDD52 pKa = 4.48LDD54 pKa = 4.3LAPFVPLDD62 pKa = 3.35NYY64 pKa = 10.85VDD66 pKa = 4.61FGYY69 pKa = 10.96EE70 pKa = 3.74SDD72 pKa = 4.3EE73 pKa = 4.5DD74 pKa = 4.25FGLQTVPNLPPSVLLFCDD92 pKa = 4.27APASFPDD99 pKa = 3.33RR100 pKa = 11.84TEE102 pKa = 4.15IDD104 pKa = 3.45LHH106 pKa = 8.45CYY108 pKa = 10.27EE109 pKa = 5.02SMPSSVEE116 pKa = 3.79STGAISPPPSAQVSSSPSDD135 pKa = 3.8CDD137 pKa = 4.02QHH139 pKa = 8.18ICDD142 pKa = 5.26PGNSPCSLCFLRR154 pKa = 11.84LAYY157 pKa = 10.57SGMCFSYY164 pKa = 10.2TFSHH168 pKa = 7.15LLCC171 pKa = 5.17
Molecular weight: 18.63 kDa
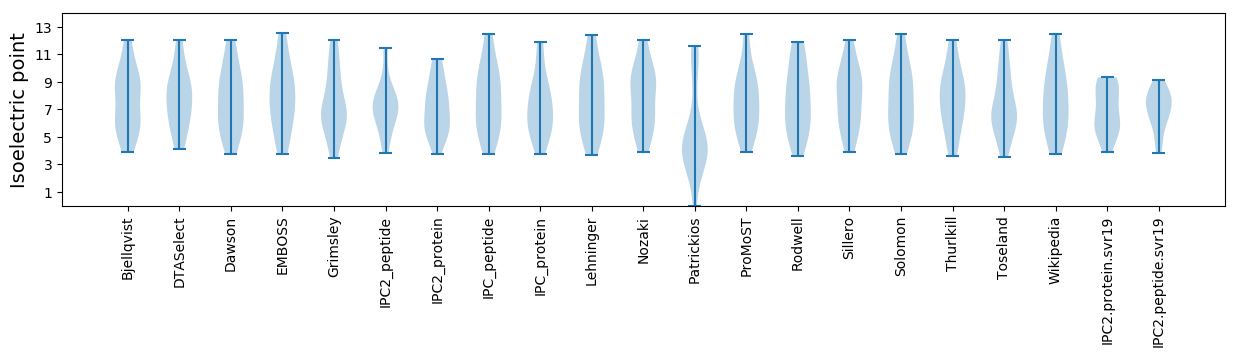
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A344X9U9|A0A344X9U9_9ADEN PX OS=Rousettus aegyptiacus adenovirus OX=1967296 PE=4 SV=1
MM1 pKa = 8.17DD2 pKa = 5.38NLTNPSRR9 pKa = 11.84LIKK12 pKa = 10.68EE13 pKa = 4.17EE14 pKa = 3.83MLQTFVPEE22 pKa = 4.09VYY24 pKa = 10.48GPEE27 pKa = 3.8RR28 pKa = 11.84VGKK31 pKa = 10.19VGRR34 pKa = 11.84KK35 pKa = 8.45RR36 pKa = 11.84KK37 pKa = 9.49RR38 pKa = 11.84DD39 pKa = 3.48NPYY42 pKa = 9.62VSLGSRR48 pKa = 11.84RR49 pKa = 11.84VAFNSPYY56 pKa = 11.09AMPVKK61 pKa = 10.48DD62 pKa = 4.33MPRR65 pKa = 11.84GAYY68 pKa = 6.92QWRR71 pKa = 11.84GRR73 pKa = 11.84RR74 pKa = 11.84VKK76 pKa = 10.53RR77 pKa = 11.84IVRR80 pKa = 11.84PGTVIVQNNRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84ATKK96 pKa = 10.08RR97 pKa = 11.84SSDD100 pKa = 3.83EE101 pKa = 3.66IFADD105 pKa = 4.57EE106 pKa = 6.36DD107 pKa = 3.83ILTQAANRR115 pKa = 11.84EE116 pKa = 4.15GEE118 pKa = 3.94FAYY121 pKa = 10.11GKK123 pKa = 9.59RR124 pKa = 11.84AHH126 pKa = 6.82IVQQGSKK133 pKa = 9.53RR134 pKa = 11.84KK135 pKa = 9.3RR136 pKa = 11.84YY137 pKa = 9.43DD138 pKa = 3.19VEE140 pKa = 4.1EE141 pKa = 4.46EE142 pKa = 4.02EE143 pKa = 4.26SSKK146 pKa = 10.99RR147 pKa = 11.84MRR149 pKa = 11.84TGDD152 pKa = 4.65FITEE156 pKa = 4.07TLKK159 pKa = 10.89RR160 pKa = 11.84QAEE163 pKa = 4.18EE164 pKa = 4.68EE165 pKa = 3.99IAQRR169 pKa = 11.84PIRR172 pKa = 11.84RR173 pKa = 11.84AVKK176 pKa = 8.84RR177 pKa = 11.84RR178 pKa = 11.84VVEE181 pKa = 4.09EE182 pKa = 3.75VDD184 pKa = 2.8AGPRR188 pKa = 11.84KK189 pKa = 9.3RR190 pKa = 11.84AYY192 pKa = 10.07VALDD196 pKa = 3.39TDD198 pKa = 3.87NPTPALEE205 pKa = 4.4AVTEE209 pKa = 4.04QAIVPMNTGDD219 pKa = 4.02EE220 pKa = 4.37MVQATAQVLQTKK232 pKa = 9.84RR233 pKa = 11.84GRR235 pKa = 11.84VTEE238 pKa = 3.94RR239 pKa = 11.84SEE241 pKa = 3.96RR242 pKa = 11.84AEE244 pKa = 4.01SNIPVEE250 pKa = 4.31VPIAEE255 pKa = 4.11GRR257 pKa = 11.84RR258 pKa = 11.84VRR260 pKa = 11.84GALKK264 pKa = 9.76VRR266 pKa = 11.84PIKK269 pKa = 10.39RR270 pKa = 11.84VAPGIGVRR278 pKa = 11.84TVDD281 pKa = 3.73LEE283 pKa = 4.03IPLRR287 pKa = 11.84EE288 pKa = 4.12SVTNVSIPSEE298 pKa = 4.26YY299 pKa = 10.58EE300 pKa = 3.72RR301 pKa = 11.84MEE303 pKa = 4.16TTVTRR308 pKa = 11.84SVRR311 pKa = 11.84RR312 pKa = 11.84QPVRR316 pKa = 11.84RR317 pKa = 11.84TQKK320 pKa = 8.76TYY322 pKa = 10.9KK323 pKa = 8.64PANSIIPAAVYY334 pKa = 10.0HH335 pKa = 6.46PSIQHH340 pKa = 5.9PRR342 pKa = 11.84ALPKK346 pKa = 9.88KK347 pKa = 8.47EE348 pKa = 4.07RR349 pKa = 11.84IIPVVRR355 pKa = 11.84YY356 pKa = 8.91HH357 pKa = 6.87PSITMPRR364 pKa = 11.84RR365 pKa = 11.84YY366 pKa = 10.05
MM1 pKa = 8.17DD2 pKa = 5.38NLTNPSRR9 pKa = 11.84LIKK12 pKa = 10.68EE13 pKa = 4.17EE14 pKa = 3.83MLQTFVPEE22 pKa = 4.09VYY24 pKa = 10.48GPEE27 pKa = 3.8RR28 pKa = 11.84VGKK31 pKa = 10.19VGRR34 pKa = 11.84KK35 pKa = 8.45RR36 pKa = 11.84KK37 pKa = 9.49RR38 pKa = 11.84DD39 pKa = 3.48NPYY42 pKa = 9.62VSLGSRR48 pKa = 11.84RR49 pKa = 11.84VAFNSPYY56 pKa = 11.09AMPVKK61 pKa = 10.48DD62 pKa = 4.33MPRR65 pKa = 11.84GAYY68 pKa = 6.92QWRR71 pKa = 11.84GRR73 pKa = 11.84RR74 pKa = 11.84VKK76 pKa = 10.53RR77 pKa = 11.84IVRR80 pKa = 11.84PGTVIVQNNRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84ATKK96 pKa = 10.08RR97 pKa = 11.84SSDD100 pKa = 3.83EE101 pKa = 3.66IFADD105 pKa = 4.57EE106 pKa = 6.36DD107 pKa = 3.83ILTQAANRR115 pKa = 11.84EE116 pKa = 4.15GEE118 pKa = 3.94FAYY121 pKa = 10.11GKK123 pKa = 9.59RR124 pKa = 11.84AHH126 pKa = 6.82IVQQGSKK133 pKa = 9.53RR134 pKa = 11.84KK135 pKa = 9.3RR136 pKa = 11.84YY137 pKa = 9.43DD138 pKa = 3.19VEE140 pKa = 4.1EE141 pKa = 4.46EE142 pKa = 4.02EE143 pKa = 4.26SSKK146 pKa = 10.99RR147 pKa = 11.84MRR149 pKa = 11.84TGDD152 pKa = 4.65FITEE156 pKa = 4.07TLKK159 pKa = 10.89RR160 pKa = 11.84QAEE163 pKa = 4.18EE164 pKa = 4.68EE165 pKa = 3.99IAQRR169 pKa = 11.84PIRR172 pKa = 11.84RR173 pKa = 11.84AVKK176 pKa = 8.84RR177 pKa = 11.84RR178 pKa = 11.84VVEE181 pKa = 4.09EE182 pKa = 3.75VDD184 pKa = 2.8AGPRR188 pKa = 11.84KK189 pKa = 9.3RR190 pKa = 11.84AYY192 pKa = 10.07VALDD196 pKa = 3.39TDD198 pKa = 3.87NPTPALEE205 pKa = 4.4AVTEE209 pKa = 4.04QAIVPMNTGDD219 pKa = 4.02EE220 pKa = 4.37MVQATAQVLQTKK232 pKa = 9.84RR233 pKa = 11.84GRR235 pKa = 11.84VTEE238 pKa = 3.94RR239 pKa = 11.84SEE241 pKa = 3.96RR242 pKa = 11.84AEE244 pKa = 4.01SNIPVEE250 pKa = 4.31VPIAEE255 pKa = 4.11GRR257 pKa = 11.84RR258 pKa = 11.84VRR260 pKa = 11.84GALKK264 pKa = 9.76VRR266 pKa = 11.84PIKK269 pKa = 10.39RR270 pKa = 11.84VAPGIGVRR278 pKa = 11.84TVDD281 pKa = 3.73LEE283 pKa = 4.03IPLRR287 pKa = 11.84EE288 pKa = 4.12SVTNVSIPSEE298 pKa = 4.26YY299 pKa = 10.58EE300 pKa = 3.72RR301 pKa = 11.84MEE303 pKa = 4.16TTVTRR308 pKa = 11.84SVRR311 pKa = 11.84RR312 pKa = 11.84QPVRR316 pKa = 11.84RR317 pKa = 11.84TQKK320 pKa = 8.76TYY322 pKa = 10.9KK323 pKa = 8.64PANSIIPAAVYY334 pKa = 10.0HH335 pKa = 6.46PSIQHH340 pKa = 5.9PRR342 pKa = 11.84ALPKK346 pKa = 9.88KK347 pKa = 8.47EE348 pKa = 4.07RR349 pKa = 11.84IIPVVRR355 pKa = 11.84YY356 pKa = 8.91HH357 pKa = 6.87PSITMPRR364 pKa = 11.84RR365 pKa = 11.84YY366 pKa = 10.05
Molecular weight: 41.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8185 |
55 |
973 |
327.4 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.097 ± 0.254 | 1.698 ± 0.32 |
4.52 ± 0.309 | 6.158 ± 0.461 |
4.704 ± 0.297 | 5.437 ± 0.475 |
1.894 ± 0.141 | 5.767 ± 0.32 |
6.133 ± 0.635 | 9.42 ± 0.494 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.346 ± 0.146 | 6.182 ± 0.491 |
5.816 ± 0.296 | 4.301 ± 0.352 |
5.18 ± 0.77 | 7.221 ± 0.393 |
5.999 ± 0.344 | 5.999 ± 0.261 |
1.1 ± 0.148 | 4.032 ± 0.392 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
