
Cordyline virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Velarivirus
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
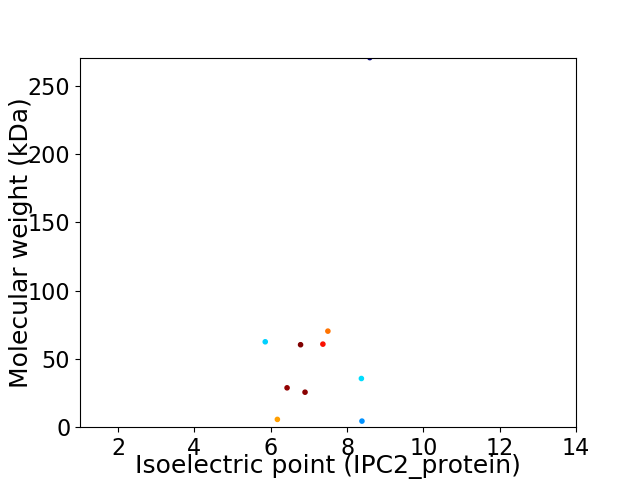
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7CT66|E7CT66_9CLOS p6 OS=Cordyline virus 1 OX=937809 PE=4 SV=1
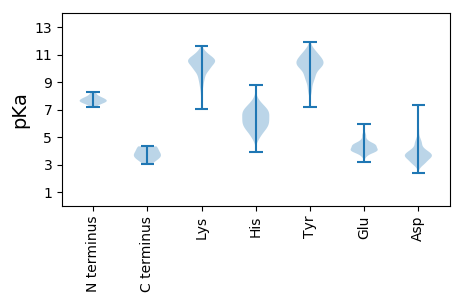
MM1 pKa = 7.55PTFCRR6 pKa = 11.84AGVDD10 pKa = 4.04FGTTFSTISAFVDD23 pKa = 4.48EE24 pKa = 4.81VATPLFLEE32 pKa = 5.23GSPFIPTVITFFDD45 pKa = 3.56NKK47 pKa = 10.05VVIGEE52 pKa = 4.08LAKK55 pKa = 10.27TISEE59 pKa = 4.35VVKK62 pKa = 11.17DD63 pKa = 3.56NVTYY67 pKa = 10.71FDD69 pKa = 3.73LKK71 pKa = 10.3RR72 pKa = 11.84WVGVNEE78 pKa = 4.0KK79 pKa = 10.9NFIKK83 pKa = 10.48LKK85 pKa = 10.58EE86 pKa = 4.04KK87 pKa = 10.42LQPSYY92 pKa = 10.83EE93 pKa = 4.25CKK95 pKa = 10.43LEE97 pKa = 4.36KK98 pKa = 10.75GEE100 pKa = 4.72CYY102 pKa = 9.54MGGIGVRR109 pKa = 11.84TVFRR113 pKa = 11.84SVGFLIATYY122 pKa = 10.9LDD124 pKa = 3.78VLTKK128 pKa = 10.71LFEE131 pKa = 4.16EE132 pKa = 5.09KK133 pKa = 10.77YY134 pKa = 10.32DD135 pKa = 4.01VKK137 pKa = 10.19ITNLNVSVPADD148 pKa = 4.42FYY150 pKa = 10.3TFQRR154 pKa = 11.84SYY156 pKa = 10.32MRR158 pKa = 11.84NAVNNLGIKK167 pKa = 8.8VDD169 pKa = 3.6RR170 pKa = 11.84MINEE174 pKa = 4.14PSAAALHH181 pKa = 6.58SILSNPEE188 pKa = 3.27FTDD191 pKa = 4.15FVIFDD196 pKa = 4.08FGGGTFDD203 pKa = 3.81VSYY206 pKa = 10.39IKK208 pKa = 10.71KK209 pKa = 9.07KK210 pKa = 10.75GKK212 pKa = 10.1IIMICDD218 pKa = 3.59TQGDD222 pKa = 4.46LFLGGRR228 pKa = 11.84DD229 pKa = 3.02IDD231 pKa = 4.91KK232 pKa = 10.93SIQHH236 pKa = 5.75YY237 pKa = 10.07FFGKK241 pKa = 10.05YY242 pKa = 9.99SIEE245 pKa = 3.6IHH247 pKa = 6.33PFSLSYY253 pKa = 10.0MKK255 pKa = 10.68EE256 pKa = 3.91GVSTGKK262 pKa = 9.69SQSFNVLSSNKK273 pKa = 9.66EE274 pKa = 3.69INHH277 pKa = 5.44VNFSKK282 pKa = 11.22DD283 pKa = 3.37EE284 pKa = 4.32LNSIVSPFAKK294 pKa = 9.96RR295 pKa = 11.84SCEE298 pKa = 3.62ILKK301 pKa = 10.74AVIDD305 pKa = 4.04RR306 pKa = 11.84NEE308 pKa = 3.6ITNAVICMVGGSSLLTEE325 pKa = 5.07VYY327 pKa = 10.64NQVNNVAKK335 pKa = 9.24GTNNKK340 pKa = 8.95IFRR343 pKa = 11.84DD344 pKa = 3.55EE345 pKa = 3.94NLRR348 pKa = 11.84LSVSFGCSCLHH359 pKa = 6.92FFTDD363 pKa = 5.52DD364 pKa = 3.62PDD366 pKa = 3.48FTYY369 pKa = 10.61IDD371 pKa = 4.09VNSHH375 pKa = 5.79CVFEE379 pKa = 5.0IDD381 pKa = 4.7EE382 pKa = 4.24MFKK385 pKa = 10.75PSVLIRR391 pKa = 11.84KK392 pKa = 7.67PMPIPYY398 pKa = 8.97TLRR401 pKa = 11.84QEE403 pKa = 4.21KK404 pKa = 10.27QNNNKK409 pKa = 9.5FLTAVDD415 pKa = 3.67IYY417 pKa = 10.91EE418 pKa = 5.14GDD420 pKa = 3.93SQWFLDD426 pKa = 4.12CQVLIKK432 pKa = 10.56DD433 pKa = 4.18VYY435 pKa = 9.84STDD438 pKa = 3.49AVSSLGSGYY447 pKa = 10.61YY448 pKa = 9.36RR449 pKa = 11.84VIQYY453 pKa = 11.09DD454 pKa = 3.49LDD456 pKa = 3.94GNINVWIEE464 pKa = 4.11NKK466 pKa = 10.54DD467 pKa = 3.57STNKK471 pKa = 10.05RR472 pKa = 11.84NLKK475 pKa = 10.45SLITSHH481 pKa = 6.18KK482 pKa = 8.79TIKK485 pKa = 10.62LEE487 pKa = 3.76NFEE490 pKa = 4.39RR491 pKa = 11.84VQTGSSSLYY500 pKa = 8.67CTVAEE505 pKa = 4.45LIKK508 pKa = 10.43YY509 pKa = 9.3HH510 pKa = 6.96KK511 pKa = 10.09LDD513 pKa = 3.92KK514 pKa = 10.96NIEE517 pKa = 4.53AIDD520 pKa = 3.66SNLFFKK526 pKa = 10.71IKK528 pKa = 10.37DD529 pKa = 3.66YY530 pKa = 11.5IEE532 pKa = 4.4INGGISKK539 pKa = 10.67YY540 pKa = 10.39IEE542 pKa = 3.96TLRR545 pKa = 11.84KK546 pKa = 10.14NGVQII551 pKa = 4.32
MM1 pKa = 7.55PTFCRR6 pKa = 11.84AGVDD10 pKa = 4.04FGTTFSTISAFVDD23 pKa = 4.48EE24 pKa = 4.81VATPLFLEE32 pKa = 5.23GSPFIPTVITFFDD45 pKa = 3.56NKK47 pKa = 10.05VVIGEE52 pKa = 4.08LAKK55 pKa = 10.27TISEE59 pKa = 4.35VVKK62 pKa = 11.17DD63 pKa = 3.56NVTYY67 pKa = 10.71FDD69 pKa = 3.73LKK71 pKa = 10.3RR72 pKa = 11.84WVGVNEE78 pKa = 4.0KK79 pKa = 10.9NFIKK83 pKa = 10.48LKK85 pKa = 10.58EE86 pKa = 4.04KK87 pKa = 10.42LQPSYY92 pKa = 10.83EE93 pKa = 4.25CKK95 pKa = 10.43LEE97 pKa = 4.36KK98 pKa = 10.75GEE100 pKa = 4.72CYY102 pKa = 9.54MGGIGVRR109 pKa = 11.84TVFRR113 pKa = 11.84SVGFLIATYY122 pKa = 10.9LDD124 pKa = 3.78VLTKK128 pKa = 10.71LFEE131 pKa = 4.16EE132 pKa = 5.09KK133 pKa = 10.77YY134 pKa = 10.32DD135 pKa = 4.01VKK137 pKa = 10.19ITNLNVSVPADD148 pKa = 4.42FYY150 pKa = 10.3TFQRR154 pKa = 11.84SYY156 pKa = 10.32MRR158 pKa = 11.84NAVNNLGIKK167 pKa = 8.8VDD169 pKa = 3.6RR170 pKa = 11.84MINEE174 pKa = 4.14PSAAALHH181 pKa = 6.58SILSNPEE188 pKa = 3.27FTDD191 pKa = 4.15FVIFDD196 pKa = 4.08FGGGTFDD203 pKa = 3.81VSYY206 pKa = 10.39IKK208 pKa = 10.71KK209 pKa = 9.07KK210 pKa = 10.75GKK212 pKa = 10.1IIMICDD218 pKa = 3.59TQGDD222 pKa = 4.46LFLGGRR228 pKa = 11.84DD229 pKa = 3.02IDD231 pKa = 4.91KK232 pKa = 10.93SIQHH236 pKa = 5.75YY237 pKa = 10.07FFGKK241 pKa = 10.05YY242 pKa = 9.99SIEE245 pKa = 3.6IHH247 pKa = 6.33PFSLSYY253 pKa = 10.0MKK255 pKa = 10.68EE256 pKa = 3.91GVSTGKK262 pKa = 9.69SQSFNVLSSNKK273 pKa = 9.66EE274 pKa = 3.69INHH277 pKa = 5.44VNFSKK282 pKa = 11.22DD283 pKa = 3.37EE284 pKa = 4.32LNSIVSPFAKK294 pKa = 9.96RR295 pKa = 11.84SCEE298 pKa = 3.62ILKK301 pKa = 10.74AVIDD305 pKa = 4.04RR306 pKa = 11.84NEE308 pKa = 3.6ITNAVICMVGGSSLLTEE325 pKa = 5.07VYY327 pKa = 10.64NQVNNVAKK335 pKa = 9.24GTNNKK340 pKa = 8.95IFRR343 pKa = 11.84DD344 pKa = 3.55EE345 pKa = 3.94NLRR348 pKa = 11.84LSVSFGCSCLHH359 pKa = 6.92FFTDD363 pKa = 5.52DD364 pKa = 3.62PDD366 pKa = 3.48FTYY369 pKa = 10.61IDD371 pKa = 4.09VNSHH375 pKa = 5.79CVFEE379 pKa = 5.0IDD381 pKa = 4.7EE382 pKa = 4.24MFKK385 pKa = 10.75PSVLIRR391 pKa = 11.84KK392 pKa = 7.67PMPIPYY398 pKa = 8.97TLRR401 pKa = 11.84QEE403 pKa = 4.21KK404 pKa = 10.27QNNNKK409 pKa = 9.5FLTAVDD415 pKa = 3.67IYY417 pKa = 10.91EE418 pKa = 5.14GDD420 pKa = 3.93SQWFLDD426 pKa = 4.12CQVLIKK432 pKa = 10.56DD433 pKa = 4.18VYY435 pKa = 9.84STDD438 pKa = 3.49AVSSLGSGYY447 pKa = 10.61YY448 pKa = 9.36RR449 pKa = 11.84VIQYY453 pKa = 11.09DD454 pKa = 3.49LDD456 pKa = 3.94GNINVWIEE464 pKa = 4.11NKK466 pKa = 10.54DD467 pKa = 3.57STNKK471 pKa = 10.05RR472 pKa = 11.84NLKK475 pKa = 10.45SLITSHH481 pKa = 6.18KK482 pKa = 8.79TIKK485 pKa = 10.62LEE487 pKa = 3.76NFEE490 pKa = 4.39RR491 pKa = 11.84VQTGSSSLYY500 pKa = 8.67CTVAEE505 pKa = 4.45LIKK508 pKa = 10.43YY509 pKa = 9.3HH510 pKa = 6.96KK511 pKa = 10.09LDD513 pKa = 3.92KK514 pKa = 10.96NIEE517 pKa = 4.53AIDD520 pKa = 3.66SNLFFKK526 pKa = 10.71IKK528 pKa = 10.37DD529 pKa = 3.66YY530 pKa = 11.5IEE532 pKa = 4.4INGGISKK539 pKa = 10.67YY540 pKa = 10.39IEE542 pKa = 3.96TLRR545 pKa = 11.84KK546 pKa = 10.14NGVQII551 pKa = 4.32
Molecular weight: 62.53 kDa
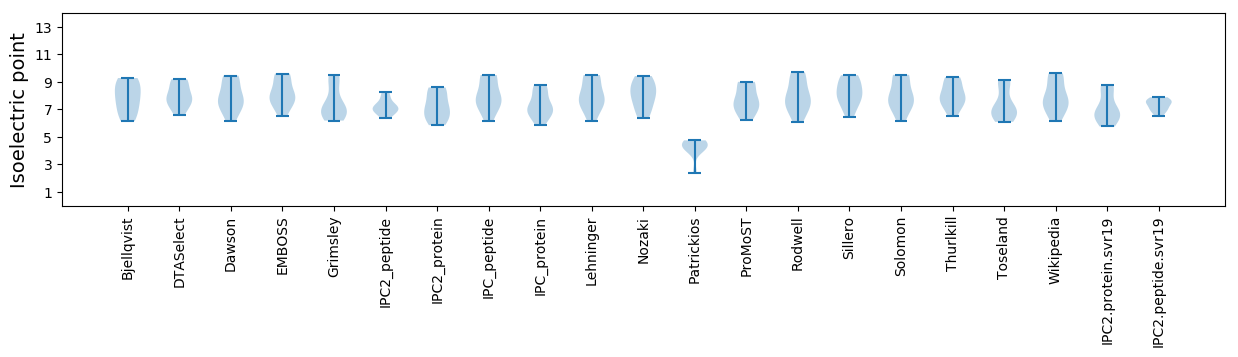
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7CT65|E7CT65_9CLOS HSP70h OS=Cordyline virus 1 OX=937809 PE=3 SV=1
MM1 pKa = 7.58IFLFFLFFLLFLFLLNLCTSPSNKK25 pKa = 9.6GFIVHH30 pKa = 5.68KK31 pKa = 10.08QIYY34 pKa = 8.52YY35 pKa = 10.52SPP37 pKa = 3.86
MM1 pKa = 7.58IFLFFLFFLLFLFLLNLCTSPSNKK25 pKa = 9.6GFIVHH30 pKa = 5.68KK31 pKa = 10.08QIYY34 pKa = 8.52YY35 pKa = 10.52SPP37 pKa = 3.86
Molecular weight: 4.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5415 |
37 |
2360 |
541.5 |
62.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.358 ± 0.413 | 1.773 ± 0.229 |
5.854 ± 0.281 | 5.669 ± 0.558 |
6.464 ± 0.431 | 3.767 ± 0.359 |
1.699 ± 0.179 | 7.701 ± 0.591 |
8.79 ± 0.398 | 9.03 ± 0.331 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.161 ± 0.274 | 7.313 ± 0.298 |
2.77 ± 0.363 | 2.77 ± 0.285 |
4.543 ± 0.285 | 7.553 ± 0.374 |
5.706 ± 0.586 | 7.018 ± 0.538 |
0.517 ± 0.109 | 4.543 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
