
Collinsella sp. An271
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; unclassified Collinsella
Average proteome isoelectric point is 5.73
Get precalculated fractions of proteins
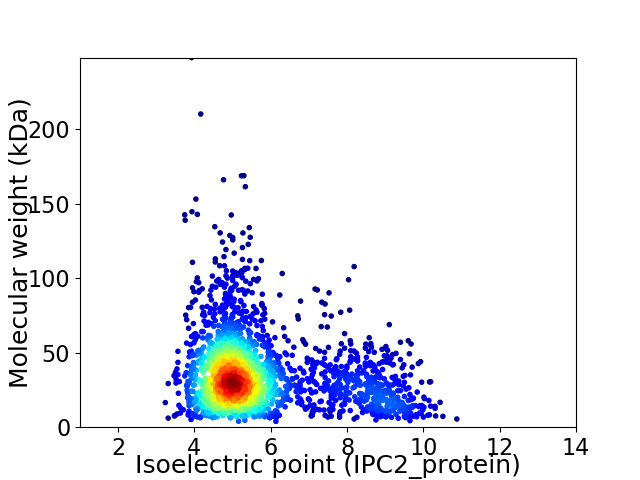
Virtual 2D-PAGE plot for 2335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
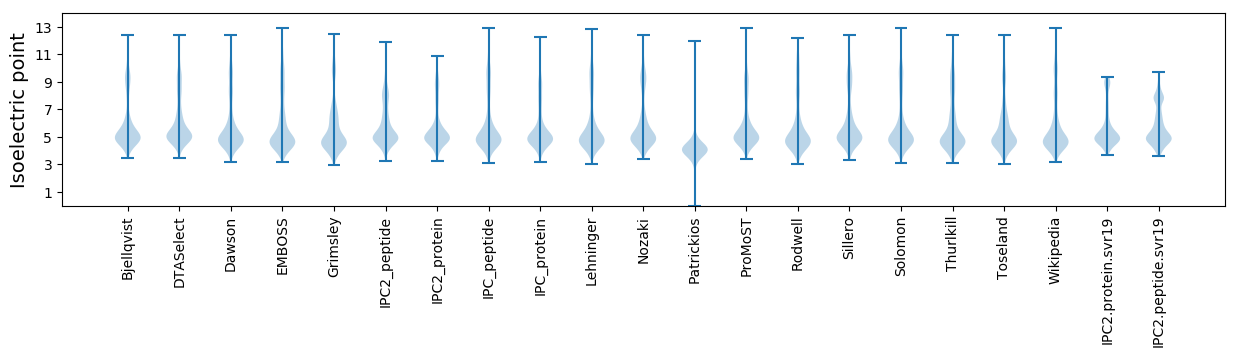
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4DG65|A0A1Y4DG65_9ACTN DNA primase OS=Collinsella sp. An271 OX=1965616 GN=dnaG PE=3 SV=1
MM1 pKa = 7.43KK2 pKa = 10.05RR3 pKa = 11.84FHH5 pKa = 6.64SVWRR9 pKa = 11.84ACALSCALVAGLSLAPIIAGATAADD34 pKa = 3.57AAAATDD40 pKa = 5.28GILITLDD47 pKa = 3.35SAASAQARR55 pKa = 11.84SLGGDD60 pKa = 3.58DD61 pKa = 3.94VNLLAEE67 pKa = 4.45SEE69 pKa = 4.72VVAQLGQAGLEE80 pKa = 4.28VNDD83 pKa = 4.14SFEE86 pKa = 4.41SQGGQVVLTADD97 pKa = 3.57TTGDD101 pKa = 3.6LSDD104 pKa = 4.53EE105 pKa = 4.21EE106 pKa = 5.0ALAQARR112 pKa = 11.84DD113 pKa = 3.55IEE115 pKa = 4.64GVADD119 pKa = 3.45VQYY122 pKa = 10.93NYY124 pKa = 10.45VYY126 pKa = 10.46HH127 pKa = 6.63LVDD130 pKa = 5.5IIEE133 pKa = 4.41EE134 pKa = 4.31DD135 pKa = 3.53ATSSTGTSRR144 pKa = 11.84SGVSTLTVSSVNDD157 pKa = 3.57PFAQISSPAADD168 pKa = 3.52NNQYY172 pKa = 9.66WLYY175 pKa = 10.89SANLDD180 pKa = 4.42DD181 pKa = 5.5AWEE184 pKa = 4.1QAQSDD189 pKa = 4.14HH190 pKa = 7.52DD191 pKa = 3.61ITIAVLDD198 pKa = 3.9SGVDD202 pKa = 3.66LDD204 pKa = 5.53HH205 pKa = 7.51EE206 pKa = 4.5DD207 pKa = 3.33LAGNILTDD215 pKa = 4.37LAWDD219 pKa = 3.68ATLEE223 pKa = 4.23QPLSACEE230 pKa = 3.79ITDD233 pKa = 3.94CAGHH237 pKa = 5.65GTMVAGVASAVANNALGIAGASYY260 pKa = 10.87NADD263 pKa = 3.89LLPIKK268 pKa = 10.73VVDD271 pKa = 3.52NEE273 pKa = 3.88EE274 pKa = 4.01RR275 pKa = 11.84QITSADD281 pKa = 3.38LVKK284 pKa = 10.58SYY286 pKa = 10.91EE287 pKa = 4.0YY288 pKa = 11.19LFDD291 pKa = 4.61LVDD294 pKa = 4.37SGRR297 pKa = 11.84LDD299 pKa = 3.48NLRR302 pKa = 11.84VINMSLGGYY311 pKa = 9.15EE312 pKa = 4.5DD313 pKa = 3.94SQANDD318 pKa = 3.21EE319 pKa = 4.65AFHH322 pKa = 6.99DD323 pKa = 4.62VIKK326 pKa = 9.62TALNTYY332 pKa = 10.42DD333 pKa = 4.06ILTVCAGGNGNQTTTPNTGKK353 pKa = 9.76MYY355 pKa = 10.07PADD358 pKa = 4.02FDD360 pKa = 3.83EE361 pKa = 5.9CISVTALEE369 pKa = 4.24SDD371 pKa = 3.59GTNLVWSDD379 pKa = 3.39YY380 pKa = 11.73NEE382 pKa = 4.38FKK384 pKa = 10.79DD385 pKa = 4.88ISAPGQNIRR394 pKa = 11.84TTNMSGGFSTASGTSVASPIVSGTVALLYY423 pKa = 10.74AADD426 pKa = 4.28PDD428 pKa = 3.85ATPEE432 pKa = 3.9QVRR435 pKa = 11.84AALEE439 pKa = 4.05EE440 pKa = 4.13TAVPVVDD447 pKa = 4.38EE448 pKa = 4.17EE449 pKa = 4.28HH450 pKa = 7.36DD451 pKa = 3.61RR452 pKa = 11.84TEE454 pKa = 4.12VSGSAGALDD463 pKa = 3.7AGAAVEE469 pKa = 4.06QLTGAVEE476 pKa = 4.22PEE478 pKa = 4.55PEE480 pKa = 4.85PDD482 pKa = 4.01PDD484 pKa = 4.18PEE486 pKa = 4.77PEE488 pKa = 4.44PEE490 pKa = 4.27LPFVDD495 pKa = 5.4VDD497 pKa = 3.9HH498 pKa = 6.69EE499 pKa = 4.53WYY501 pKa = 10.42YY502 pKa = 10.03PAIVYY507 pKa = 9.95VYY509 pKa = 9.96EE510 pKa = 3.94NDD512 pKa = 3.33IMHH515 pKa = 7.28GLSDD519 pKa = 4.08SPRR522 pKa = 11.84FAPNEE527 pKa = 4.18PIVRR531 pKa = 11.84QDD533 pKa = 3.3AAVVLYY539 pKa = 10.76NILGNGEE546 pKa = 4.15KK547 pKa = 10.66APDD550 pKa = 4.07CGLVDD555 pKa = 3.88VGQDD559 pKa = 3.53YY560 pKa = 8.18YY561 pKa = 11.44TDD563 pKa = 3.47AVNWAVAHH571 pKa = 6.43GYY573 pKa = 9.81INGYY577 pKa = 9.35EE578 pKa = 3.94NVATGEE584 pKa = 4.2VTHH587 pKa = 6.7FGVGDD592 pKa = 3.83PLTRR596 pKa = 11.84EE597 pKa = 3.69QLACIVANIARR608 pKa = 11.84NGEE611 pKa = 4.27PFGSMAKK618 pKa = 9.8YY619 pKa = 10.76EE620 pKa = 4.34SMPDD624 pKa = 3.23HH625 pKa = 7.65DD626 pKa = 5.36KK627 pKa = 10.75TSTWAVTNVVWAINNQIINGVEE649 pKa = 3.95LGDD652 pKa = 3.63VRR654 pKa = 11.84YY655 pKa = 9.44VAPQNKK661 pKa = 7.64ATRR664 pKa = 11.84AEE666 pKa = 4.03MAAIMMNCLEE676 pKa = 3.98QDD678 pKa = 3.71VFF680 pKa = 4.18
MM1 pKa = 7.43KK2 pKa = 10.05RR3 pKa = 11.84FHH5 pKa = 6.64SVWRR9 pKa = 11.84ACALSCALVAGLSLAPIIAGATAADD34 pKa = 3.57AAAATDD40 pKa = 5.28GILITLDD47 pKa = 3.35SAASAQARR55 pKa = 11.84SLGGDD60 pKa = 3.58DD61 pKa = 3.94VNLLAEE67 pKa = 4.45SEE69 pKa = 4.72VVAQLGQAGLEE80 pKa = 4.28VNDD83 pKa = 4.14SFEE86 pKa = 4.41SQGGQVVLTADD97 pKa = 3.57TTGDD101 pKa = 3.6LSDD104 pKa = 4.53EE105 pKa = 4.21EE106 pKa = 5.0ALAQARR112 pKa = 11.84DD113 pKa = 3.55IEE115 pKa = 4.64GVADD119 pKa = 3.45VQYY122 pKa = 10.93NYY124 pKa = 10.45VYY126 pKa = 10.46HH127 pKa = 6.63LVDD130 pKa = 5.5IIEE133 pKa = 4.41EE134 pKa = 4.31DD135 pKa = 3.53ATSSTGTSRR144 pKa = 11.84SGVSTLTVSSVNDD157 pKa = 3.57PFAQISSPAADD168 pKa = 3.52NNQYY172 pKa = 9.66WLYY175 pKa = 10.89SANLDD180 pKa = 4.42DD181 pKa = 5.5AWEE184 pKa = 4.1QAQSDD189 pKa = 4.14HH190 pKa = 7.52DD191 pKa = 3.61ITIAVLDD198 pKa = 3.9SGVDD202 pKa = 3.66LDD204 pKa = 5.53HH205 pKa = 7.51EE206 pKa = 4.5DD207 pKa = 3.33LAGNILTDD215 pKa = 4.37LAWDD219 pKa = 3.68ATLEE223 pKa = 4.23QPLSACEE230 pKa = 3.79ITDD233 pKa = 3.94CAGHH237 pKa = 5.65GTMVAGVASAVANNALGIAGASYY260 pKa = 10.87NADD263 pKa = 3.89LLPIKK268 pKa = 10.73VVDD271 pKa = 3.52NEE273 pKa = 3.88EE274 pKa = 4.01RR275 pKa = 11.84QITSADD281 pKa = 3.38LVKK284 pKa = 10.58SYY286 pKa = 10.91EE287 pKa = 4.0YY288 pKa = 11.19LFDD291 pKa = 4.61LVDD294 pKa = 4.37SGRR297 pKa = 11.84LDD299 pKa = 3.48NLRR302 pKa = 11.84VINMSLGGYY311 pKa = 9.15EE312 pKa = 4.5DD313 pKa = 3.94SQANDD318 pKa = 3.21EE319 pKa = 4.65AFHH322 pKa = 6.99DD323 pKa = 4.62VIKK326 pKa = 9.62TALNTYY332 pKa = 10.42DD333 pKa = 4.06ILTVCAGGNGNQTTTPNTGKK353 pKa = 9.76MYY355 pKa = 10.07PADD358 pKa = 4.02FDD360 pKa = 3.83EE361 pKa = 5.9CISVTALEE369 pKa = 4.24SDD371 pKa = 3.59GTNLVWSDD379 pKa = 3.39YY380 pKa = 11.73NEE382 pKa = 4.38FKK384 pKa = 10.79DD385 pKa = 4.88ISAPGQNIRR394 pKa = 11.84TTNMSGGFSTASGTSVASPIVSGTVALLYY423 pKa = 10.74AADD426 pKa = 4.28PDD428 pKa = 3.85ATPEE432 pKa = 3.9QVRR435 pKa = 11.84AALEE439 pKa = 4.05EE440 pKa = 4.13TAVPVVDD447 pKa = 4.38EE448 pKa = 4.17EE449 pKa = 4.28HH450 pKa = 7.36DD451 pKa = 3.61RR452 pKa = 11.84TEE454 pKa = 4.12VSGSAGALDD463 pKa = 3.7AGAAVEE469 pKa = 4.06QLTGAVEE476 pKa = 4.22PEE478 pKa = 4.55PEE480 pKa = 4.85PDD482 pKa = 4.01PDD484 pKa = 4.18PEE486 pKa = 4.77PEE488 pKa = 4.44PEE490 pKa = 4.27LPFVDD495 pKa = 5.4VDD497 pKa = 3.9HH498 pKa = 6.69EE499 pKa = 4.53WYY501 pKa = 10.42YY502 pKa = 10.03PAIVYY507 pKa = 9.95VYY509 pKa = 9.96EE510 pKa = 3.94NDD512 pKa = 3.33IMHH515 pKa = 7.28GLSDD519 pKa = 4.08SPRR522 pKa = 11.84FAPNEE527 pKa = 4.18PIVRR531 pKa = 11.84QDD533 pKa = 3.3AAVVLYY539 pKa = 10.76NILGNGEE546 pKa = 4.15KK547 pKa = 10.66APDD550 pKa = 4.07CGLVDD555 pKa = 3.88VGQDD559 pKa = 3.53YY560 pKa = 8.18YY561 pKa = 11.44TDD563 pKa = 3.47AVNWAVAHH571 pKa = 6.43GYY573 pKa = 9.81INGYY577 pKa = 9.35EE578 pKa = 3.94NVATGEE584 pKa = 4.2VTHH587 pKa = 6.7FGVGDD592 pKa = 3.83PLTRR596 pKa = 11.84EE597 pKa = 3.69QLACIVANIARR608 pKa = 11.84NGEE611 pKa = 4.27PFGSMAKK618 pKa = 9.8YY619 pKa = 10.76EE620 pKa = 4.34SMPDD624 pKa = 3.23HH625 pKa = 7.65DD626 pKa = 5.36KK627 pKa = 10.75TSTWAVTNVVWAINNQIINGVEE649 pKa = 3.95LGDD652 pKa = 3.63VRR654 pKa = 11.84YY655 pKa = 9.44VAPQNKK661 pKa = 7.64ATRR664 pKa = 11.84AEE666 pKa = 4.03MAAIMMNCLEE676 pKa = 3.98QDD678 pKa = 3.71VFF680 pKa = 4.18
Molecular weight: 72.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4DQP6|A0A1Y4DQP6_9ACTN Uncharacterized protein OS=Collinsella sp. An271 OX=1965616 GN=B5F74_04790 PE=4 SV=1
MM1 pKa = 7.41IRR3 pKa = 11.84RR4 pKa = 11.84NATKK8 pKa = 10.41RR9 pKa = 11.84NTLSSPGRR17 pKa = 11.84RR18 pKa = 11.84AAWYY22 pKa = 10.16GLLTALALCAGYY34 pKa = 10.88LEE36 pKa = 4.56VLVPLPVSVPGIKK49 pKa = 10.16LGLGNAVVLFALEE62 pKa = 4.0RR63 pKa = 11.84MGARR67 pKa = 11.84PALFIMLAKK76 pKa = 9.62VFCSMLLFSTPQMLAFSLGGGLLSWAIMAAAVRR109 pKa = 11.84LDD111 pKa = 3.63LFSEE115 pKa = 4.42VGVSVLGGVAHH126 pKa = 7.0NAGQLLVVAALLSPQVALVNAPVLAIAGVACGLAIGLILRR166 pKa = 11.84GVLAALPKK174 pKa = 10.6AGFRR178 pKa = 11.84EE179 pKa = 4.34
MM1 pKa = 7.41IRR3 pKa = 11.84RR4 pKa = 11.84NATKK8 pKa = 10.41RR9 pKa = 11.84NTLSSPGRR17 pKa = 11.84RR18 pKa = 11.84AAWYY22 pKa = 10.16GLLTALALCAGYY34 pKa = 10.88LEE36 pKa = 4.56VLVPLPVSVPGIKK49 pKa = 10.16LGLGNAVVLFALEE62 pKa = 4.0RR63 pKa = 11.84MGARR67 pKa = 11.84PALFIMLAKK76 pKa = 9.62VFCSMLLFSTPQMLAFSLGGGLLSWAIMAAAVRR109 pKa = 11.84LDD111 pKa = 3.63LFSEE115 pKa = 4.42VGVSVLGGVAHH126 pKa = 7.0NAGQLLVVAALLSPQVALVNAPVLAIAGVACGLAIGLILRR166 pKa = 11.84GVLAALPKK174 pKa = 10.6AGFRR178 pKa = 11.84EE179 pKa = 4.34
Molecular weight: 18.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
779385 |
37 |
2360 |
333.8 |
36.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.846 ± 0.081 | 1.585 ± 0.023 |
6.495 ± 0.037 | 6.568 ± 0.048 |
3.537 ± 0.031 | 8.249 ± 0.045 |
1.948 ± 0.023 | 5.3 ± 0.041 |
3.118 ± 0.042 | 9.14 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.716 ± 0.023 | 2.644 ± 0.031 |
4.357 ± 0.033 | 2.774 ± 0.03 |
6.633 ± 0.063 | 5.828 ± 0.036 |
5.337 ± 0.038 | 8.017 ± 0.043 |
1.07 ± 0.019 | 2.84 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
