
Beihai sobemo-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
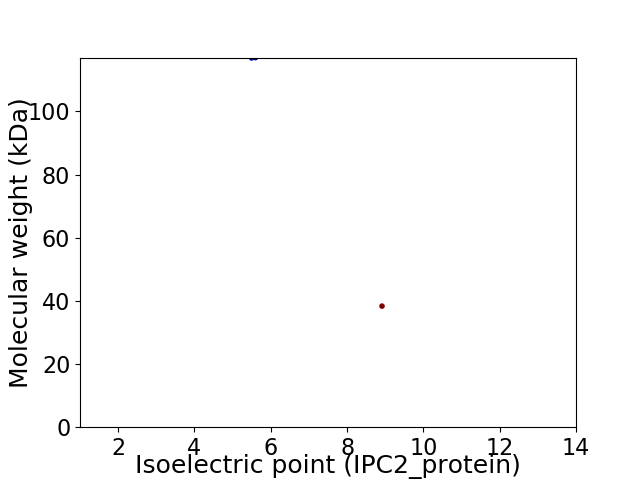
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
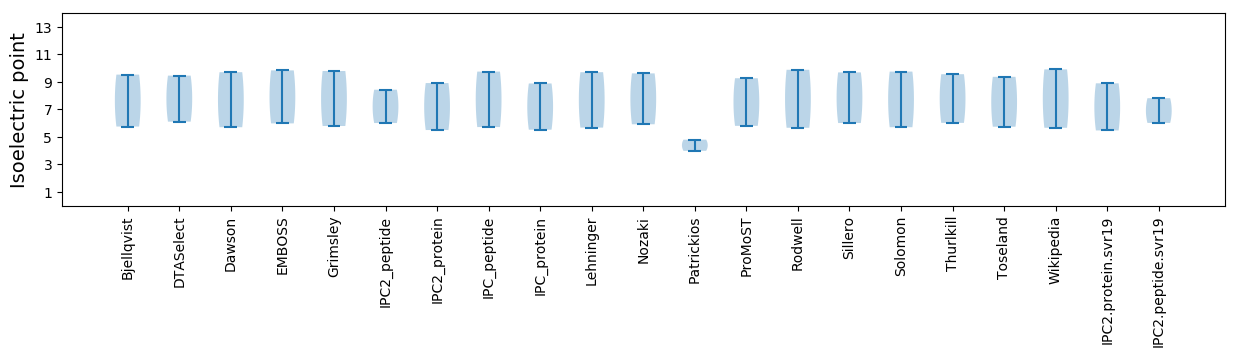
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEB0|A0A1L3KEB0_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 3 OX=1922700 PE=4 SV=1
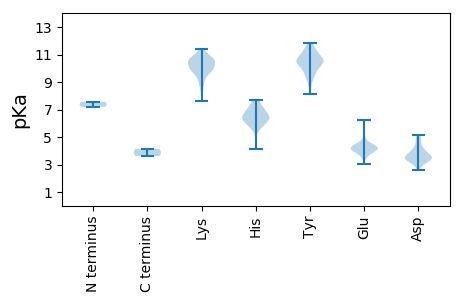
MM1 pKa = 7.57GATIAEE7 pKa = 4.36TLCIVFATLVLTRR20 pKa = 11.84LSVDD24 pKa = 3.57YY25 pKa = 10.62VTDD28 pKa = 4.29LYY30 pKa = 11.37GRR32 pKa = 11.84VYY34 pKa = 10.83SALAYY39 pKa = 8.48NAKK42 pKa = 9.91RR43 pKa = 11.84VRR45 pKa = 11.84ACMKK49 pKa = 10.58LMYY52 pKa = 10.34CPFRR56 pKa = 11.84RR57 pKa = 11.84VEE59 pKa = 4.03KK60 pKa = 10.5RR61 pKa = 11.84DD62 pKa = 3.53VFPSTEE68 pKa = 4.07GFFMHH73 pKa = 7.43PDD75 pKa = 3.15SSGVLQAYY83 pKa = 9.39YY84 pKa = 10.42RR85 pKa = 11.84FPAEE89 pKa = 3.59YY90 pKa = 10.59GGFLQRR96 pKa = 11.84LEE98 pKa = 4.36RR99 pKa = 11.84NGGYY103 pKa = 10.64GLGGEE108 pKa = 4.65YY109 pKa = 10.75VPLWKK114 pKa = 10.23EE115 pKa = 3.6AVFNKK120 pKa = 9.81VGCGEE125 pKa = 4.12SAIAGAAYY133 pKa = 9.51EE134 pKa = 4.1RR135 pKa = 11.84TVDD138 pKa = 3.39QKK140 pKa = 11.21LWKK143 pKa = 9.22GTFKK147 pKa = 11.31VNGEE151 pKa = 4.11TGVYY155 pKa = 10.68DD156 pKa = 4.55SMGLNIAGCAVITAHH171 pKa = 7.52ALLHH175 pKa = 6.22KK176 pKa = 9.89EE177 pKa = 3.97AVVLTGGNTVNVGVRR192 pKa = 11.84VEE194 pKa = 3.97TSRR197 pKa = 11.84FLKK200 pKa = 10.4PEE202 pKa = 3.45KK203 pKa = 10.11CYY205 pKa = 10.38TGSAFDD211 pKa = 3.88FAVAPLTANEE221 pKa = 3.91WSAIGAKK228 pKa = 10.12ALSSKK233 pKa = 10.71DD234 pKa = 3.58FAPVMPGPIDD244 pKa = 3.22MCFGQGVEE252 pKa = 4.09GSLFVSKK259 pKa = 10.61GQLEE263 pKa = 4.4PQVTRR268 pKa = 11.84AEE270 pKa = 4.03KK271 pKa = 10.64AGIIVASISTEE282 pKa = 3.77PGASGSVYY290 pKa = 9.7RR291 pKa = 11.84RR292 pKa = 11.84FVGGVPKK299 pKa = 9.84YY300 pKa = 9.19TGFHH304 pKa = 5.75VSRR307 pKa = 11.84PGEE310 pKa = 3.96RR311 pKa = 11.84MSQLVGKK318 pKa = 8.85YY319 pKa = 10.41NIGIDD324 pKa = 3.57FGVIFSFMRR333 pKa = 11.84KK334 pKa = 8.8NDD336 pKa = 3.45LYY338 pKa = 11.31FDD340 pKa = 3.51TTYY343 pKa = 11.36SVLRR347 pKa = 11.84KK348 pKa = 9.54LAGLSVGEE356 pKa = 4.2SFDD359 pKa = 3.79YY360 pKa = 11.35DD361 pKa = 3.43FDD363 pKa = 4.96KK364 pKa = 11.36NCKK367 pKa = 9.76EE368 pKa = 3.89PLDD371 pKa = 3.77YY372 pKa = 11.51DD373 pKa = 4.11EE374 pKa = 6.0IEE376 pKa = 4.14RR377 pKa = 11.84NYY379 pKa = 10.88LDD381 pKa = 5.12SEE383 pKa = 4.54EE384 pKa = 4.13MWQTTQDD391 pKa = 4.21EE392 pKa = 4.6ITKK395 pKa = 9.48MIYY398 pKa = 10.04GDD400 pKa = 3.8RR401 pKa = 11.84YY402 pKa = 10.86DD403 pKa = 4.04PVDD406 pKa = 3.17MHH408 pKa = 6.29VGKK411 pKa = 10.69RR412 pKa = 11.84YY413 pKa = 9.72GRR415 pKa = 11.84RR416 pKa = 11.84TRR418 pKa = 11.84DD419 pKa = 3.41QMPDD423 pKa = 3.15EE424 pKa = 4.22HH425 pKa = 7.13WYY427 pKa = 10.93GEE429 pKa = 4.37SCEE432 pKa = 4.27PPVLTVAPGLKK443 pKa = 9.53LDD445 pKa = 4.24PVIEE449 pKa = 4.38STDD452 pKa = 3.7DD453 pKa = 3.44DD454 pKa = 5.01CFDD457 pKa = 4.79DD458 pKa = 4.63ACSEE462 pKa = 4.43PGTCDD467 pKa = 3.22DD468 pKa = 4.45VAVSSPSFAEE478 pKa = 4.18DD479 pKa = 4.2DD480 pKa = 4.18EE481 pKa = 5.6LVQCSPKK488 pKa = 10.11PLSSVIARR496 pKa = 11.84AAICASAASAVALSLDD512 pKa = 3.41FSDD515 pKa = 5.01VKK517 pKa = 10.89RR518 pKa = 11.84QVLNSDD524 pKa = 4.08FSFVGKK530 pKa = 10.27LDD532 pKa = 3.74EE533 pKa = 5.53AVDD536 pKa = 3.84KK537 pKa = 11.39YY538 pKa = 11.32GADD541 pKa = 3.29AVHH544 pKa = 7.01EE545 pKa = 4.54YY546 pKa = 10.78VVSTPAFATYY556 pKa = 10.19RR557 pKa = 11.84DD558 pKa = 3.93YY559 pKa = 11.81HH560 pKa = 6.19NVTMFTEE567 pKa = 5.02TSSEE571 pKa = 3.93EE572 pKa = 4.12TLPDD576 pKa = 3.3MNGNTFFTKK585 pKa = 9.8IGEE588 pKa = 3.89YY589 pKa = 10.31RR590 pKa = 11.84IDD592 pKa = 3.75GKK594 pKa = 10.63RR595 pKa = 11.84APTTPEE601 pKa = 3.63RR602 pKa = 11.84RR603 pKa = 11.84KK604 pKa = 10.34KK605 pKa = 9.91PPKK608 pKa = 9.83NLSDD612 pKa = 3.27AAKK615 pKa = 10.24ARR617 pKa = 11.84EE618 pKa = 3.86AAVRR622 pKa = 11.84EE623 pKa = 4.29LIKK626 pKa = 10.87DD627 pKa = 3.88LSYY630 pKa = 11.44DD631 pKa = 3.46GEE633 pKa = 4.36DD634 pKa = 2.72WVTPEE639 pKa = 4.14NSRR642 pKa = 11.84KK643 pKa = 10.25NILDD647 pKa = 3.29SMRR650 pKa = 11.84AHH652 pKa = 7.24AGLACVEE659 pKa = 4.56SPPATEE665 pKa = 5.21DD666 pKa = 2.98DD667 pKa = 3.45WNRR670 pKa = 11.84ALEE673 pKa = 4.15KK674 pKa = 10.71GMEE677 pKa = 4.2VFDD680 pKa = 4.28CSPIEE685 pKa = 3.99SHH687 pKa = 6.86AEE689 pKa = 3.57RR690 pKa = 11.84GFEE693 pKa = 3.79GWYY696 pKa = 9.8KK697 pKa = 10.14QAATLQDD704 pKa = 3.42SSSGVSARR712 pKa = 11.84FRR714 pKa = 11.84SLSKK718 pKa = 10.47RR719 pKa = 11.84QWVQDD724 pKa = 3.46PQMFQVLIDD733 pKa = 3.96LVQCRR738 pKa = 11.84LVLMLIHH745 pKa = 6.55SRR747 pKa = 11.84FVQDD751 pKa = 3.5YY752 pKa = 10.39SPEE755 pKa = 4.23TVVKK759 pKa = 10.79YY760 pKa = 10.48GLKK763 pKa = 10.34DD764 pKa = 3.51VLEE767 pKa = 4.4VTVKK771 pKa = 10.66PEE773 pKa = 3.57SHH775 pKa = 6.53KK776 pKa = 10.69PEE778 pKa = 4.08KK779 pKa = 10.59AKK781 pKa = 10.6LGRR784 pKa = 11.84WRR786 pKa = 11.84LIWICSVIDD795 pKa = 3.89CLVQKK800 pKa = 10.44LLHH803 pKa = 6.3KK804 pKa = 10.44AVNARR809 pKa = 11.84DD810 pKa = 3.47IEE812 pKa = 4.21NYY814 pKa = 9.5QCGKK818 pKa = 7.65TLHH821 pKa = 6.45SAAGMGHH828 pKa = 6.63HH829 pKa = 7.7DD830 pKa = 3.83EE831 pKa = 5.81GIKK834 pKa = 9.44QLCRR838 pKa = 11.84TLKK841 pKa = 10.53EE842 pKa = 4.07LFGDD846 pKa = 3.44ATEE849 pKa = 6.25LITCDD854 pKa = 2.86ASMWDD859 pKa = 3.52FTMDD863 pKa = 3.19KK864 pKa = 10.03QAHH867 pKa = 5.59INAAKK872 pKa = 10.25RR873 pKa = 11.84RR874 pKa = 11.84VACCKK879 pKa = 10.03SEE881 pKa = 3.96PVARR885 pKa = 11.84LIMTLGHH892 pKa = 5.83VNYY895 pKa = 10.51KK896 pKa = 9.85HH897 pKa = 6.33VCEE900 pKa = 4.5CKK902 pKa = 10.24GDD904 pKa = 2.89IWRR907 pKa = 11.84CEE909 pKa = 3.85KK910 pKa = 10.9EE911 pKa = 4.25GVNGSGQCSTTSDD924 pKa = 3.28NTFSRR929 pKa = 11.84VGQARR934 pKa = 11.84ASGADD939 pKa = 3.12RR940 pKa = 11.84VIANGDD946 pKa = 3.47DD947 pKa = 3.27MAADD951 pKa = 4.26LGFDD955 pKa = 3.73PEE957 pKa = 3.99AAKK960 pKa = 10.94AFGTRR965 pKa = 11.84SRR967 pKa = 11.84DD968 pKa = 3.59VVVQDD973 pKa = 3.89ANLVPFTSHH982 pKa = 6.96HH983 pKa = 6.31INRR986 pKa = 11.84TTCTASYY993 pKa = 10.81DD994 pKa = 3.93RR995 pKa = 11.84PLKK998 pKa = 10.69LAFNLLSNCRR1008 pKa = 11.84SEE1010 pKa = 4.44DD1011 pKa = 2.95FGMRR1015 pKa = 11.84VDD1017 pKa = 3.21ALMYY1021 pKa = 9.49VVRR1024 pKa = 11.84NTPDD1028 pKa = 3.05ALAKK1032 pKa = 10.01FKK1034 pKa = 10.96KK1035 pKa = 9.97HH1036 pKa = 6.22LGPHH1040 pKa = 6.3AKK1042 pKa = 8.83GTDD1045 pKa = 3.58TCCVEE1050 pKa = 4.41MLWGAA1055 pKa = 3.62
MM1 pKa = 7.57GATIAEE7 pKa = 4.36TLCIVFATLVLTRR20 pKa = 11.84LSVDD24 pKa = 3.57YY25 pKa = 10.62VTDD28 pKa = 4.29LYY30 pKa = 11.37GRR32 pKa = 11.84VYY34 pKa = 10.83SALAYY39 pKa = 8.48NAKK42 pKa = 9.91RR43 pKa = 11.84VRR45 pKa = 11.84ACMKK49 pKa = 10.58LMYY52 pKa = 10.34CPFRR56 pKa = 11.84RR57 pKa = 11.84VEE59 pKa = 4.03KK60 pKa = 10.5RR61 pKa = 11.84DD62 pKa = 3.53VFPSTEE68 pKa = 4.07GFFMHH73 pKa = 7.43PDD75 pKa = 3.15SSGVLQAYY83 pKa = 9.39YY84 pKa = 10.42RR85 pKa = 11.84FPAEE89 pKa = 3.59YY90 pKa = 10.59GGFLQRR96 pKa = 11.84LEE98 pKa = 4.36RR99 pKa = 11.84NGGYY103 pKa = 10.64GLGGEE108 pKa = 4.65YY109 pKa = 10.75VPLWKK114 pKa = 10.23EE115 pKa = 3.6AVFNKK120 pKa = 9.81VGCGEE125 pKa = 4.12SAIAGAAYY133 pKa = 9.51EE134 pKa = 4.1RR135 pKa = 11.84TVDD138 pKa = 3.39QKK140 pKa = 11.21LWKK143 pKa = 9.22GTFKK147 pKa = 11.31VNGEE151 pKa = 4.11TGVYY155 pKa = 10.68DD156 pKa = 4.55SMGLNIAGCAVITAHH171 pKa = 7.52ALLHH175 pKa = 6.22KK176 pKa = 9.89EE177 pKa = 3.97AVVLTGGNTVNVGVRR192 pKa = 11.84VEE194 pKa = 3.97TSRR197 pKa = 11.84FLKK200 pKa = 10.4PEE202 pKa = 3.45KK203 pKa = 10.11CYY205 pKa = 10.38TGSAFDD211 pKa = 3.88FAVAPLTANEE221 pKa = 3.91WSAIGAKK228 pKa = 10.12ALSSKK233 pKa = 10.71DD234 pKa = 3.58FAPVMPGPIDD244 pKa = 3.22MCFGQGVEE252 pKa = 4.09GSLFVSKK259 pKa = 10.61GQLEE263 pKa = 4.4PQVTRR268 pKa = 11.84AEE270 pKa = 4.03KK271 pKa = 10.64AGIIVASISTEE282 pKa = 3.77PGASGSVYY290 pKa = 9.7RR291 pKa = 11.84RR292 pKa = 11.84FVGGVPKK299 pKa = 9.84YY300 pKa = 9.19TGFHH304 pKa = 5.75VSRR307 pKa = 11.84PGEE310 pKa = 3.96RR311 pKa = 11.84MSQLVGKK318 pKa = 8.85YY319 pKa = 10.41NIGIDD324 pKa = 3.57FGVIFSFMRR333 pKa = 11.84KK334 pKa = 8.8NDD336 pKa = 3.45LYY338 pKa = 11.31FDD340 pKa = 3.51TTYY343 pKa = 11.36SVLRR347 pKa = 11.84KK348 pKa = 9.54LAGLSVGEE356 pKa = 4.2SFDD359 pKa = 3.79YY360 pKa = 11.35DD361 pKa = 3.43FDD363 pKa = 4.96KK364 pKa = 11.36NCKK367 pKa = 9.76EE368 pKa = 3.89PLDD371 pKa = 3.77YY372 pKa = 11.51DD373 pKa = 4.11EE374 pKa = 6.0IEE376 pKa = 4.14RR377 pKa = 11.84NYY379 pKa = 10.88LDD381 pKa = 5.12SEE383 pKa = 4.54EE384 pKa = 4.13MWQTTQDD391 pKa = 4.21EE392 pKa = 4.6ITKK395 pKa = 9.48MIYY398 pKa = 10.04GDD400 pKa = 3.8RR401 pKa = 11.84YY402 pKa = 10.86DD403 pKa = 4.04PVDD406 pKa = 3.17MHH408 pKa = 6.29VGKK411 pKa = 10.69RR412 pKa = 11.84YY413 pKa = 9.72GRR415 pKa = 11.84RR416 pKa = 11.84TRR418 pKa = 11.84DD419 pKa = 3.41QMPDD423 pKa = 3.15EE424 pKa = 4.22HH425 pKa = 7.13WYY427 pKa = 10.93GEE429 pKa = 4.37SCEE432 pKa = 4.27PPVLTVAPGLKK443 pKa = 9.53LDD445 pKa = 4.24PVIEE449 pKa = 4.38STDD452 pKa = 3.7DD453 pKa = 3.44DD454 pKa = 5.01CFDD457 pKa = 4.79DD458 pKa = 4.63ACSEE462 pKa = 4.43PGTCDD467 pKa = 3.22DD468 pKa = 4.45VAVSSPSFAEE478 pKa = 4.18DD479 pKa = 4.2DD480 pKa = 4.18EE481 pKa = 5.6LVQCSPKK488 pKa = 10.11PLSSVIARR496 pKa = 11.84AAICASAASAVALSLDD512 pKa = 3.41FSDD515 pKa = 5.01VKK517 pKa = 10.89RR518 pKa = 11.84QVLNSDD524 pKa = 4.08FSFVGKK530 pKa = 10.27LDD532 pKa = 3.74EE533 pKa = 5.53AVDD536 pKa = 3.84KK537 pKa = 11.39YY538 pKa = 11.32GADD541 pKa = 3.29AVHH544 pKa = 7.01EE545 pKa = 4.54YY546 pKa = 10.78VVSTPAFATYY556 pKa = 10.19RR557 pKa = 11.84DD558 pKa = 3.93YY559 pKa = 11.81HH560 pKa = 6.19NVTMFTEE567 pKa = 5.02TSSEE571 pKa = 3.93EE572 pKa = 4.12TLPDD576 pKa = 3.3MNGNTFFTKK585 pKa = 9.8IGEE588 pKa = 3.89YY589 pKa = 10.31RR590 pKa = 11.84IDD592 pKa = 3.75GKK594 pKa = 10.63RR595 pKa = 11.84APTTPEE601 pKa = 3.63RR602 pKa = 11.84RR603 pKa = 11.84KK604 pKa = 10.34KK605 pKa = 9.91PPKK608 pKa = 9.83NLSDD612 pKa = 3.27AAKK615 pKa = 10.24ARR617 pKa = 11.84EE618 pKa = 3.86AAVRR622 pKa = 11.84EE623 pKa = 4.29LIKK626 pKa = 10.87DD627 pKa = 3.88LSYY630 pKa = 11.44DD631 pKa = 3.46GEE633 pKa = 4.36DD634 pKa = 2.72WVTPEE639 pKa = 4.14NSRR642 pKa = 11.84KK643 pKa = 10.25NILDD647 pKa = 3.29SMRR650 pKa = 11.84AHH652 pKa = 7.24AGLACVEE659 pKa = 4.56SPPATEE665 pKa = 5.21DD666 pKa = 2.98DD667 pKa = 3.45WNRR670 pKa = 11.84ALEE673 pKa = 4.15KK674 pKa = 10.71GMEE677 pKa = 4.2VFDD680 pKa = 4.28CSPIEE685 pKa = 3.99SHH687 pKa = 6.86AEE689 pKa = 3.57RR690 pKa = 11.84GFEE693 pKa = 3.79GWYY696 pKa = 9.8KK697 pKa = 10.14QAATLQDD704 pKa = 3.42SSSGVSARR712 pKa = 11.84FRR714 pKa = 11.84SLSKK718 pKa = 10.47RR719 pKa = 11.84QWVQDD724 pKa = 3.46PQMFQVLIDD733 pKa = 3.96LVQCRR738 pKa = 11.84LVLMLIHH745 pKa = 6.55SRR747 pKa = 11.84FVQDD751 pKa = 3.5YY752 pKa = 10.39SPEE755 pKa = 4.23TVVKK759 pKa = 10.79YY760 pKa = 10.48GLKK763 pKa = 10.34DD764 pKa = 3.51VLEE767 pKa = 4.4VTVKK771 pKa = 10.66PEE773 pKa = 3.57SHH775 pKa = 6.53KK776 pKa = 10.69PEE778 pKa = 4.08KK779 pKa = 10.59AKK781 pKa = 10.6LGRR784 pKa = 11.84WRR786 pKa = 11.84LIWICSVIDD795 pKa = 3.89CLVQKK800 pKa = 10.44LLHH803 pKa = 6.3KK804 pKa = 10.44AVNARR809 pKa = 11.84DD810 pKa = 3.47IEE812 pKa = 4.21NYY814 pKa = 9.5QCGKK818 pKa = 7.65TLHH821 pKa = 6.45SAAGMGHH828 pKa = 6.63HH829 pKa = 7.7DD830 pKa = 3.83EE831 pKa = 5.81GIKK834 pKa = 9.44QLCRR838 pKa = 11.84TLKK841 pKa = 10.53EE842 pKa = 4.07LFGDD846 pKa = 3.44ATEE849 pKa = 6.25LITCDD854 pKa = 2.86ASMWDD859 pKa = 3.52FTMDD863 pKa = 3.19KK864 pKa = 10.03QAHH867 pKa = 5.59INAAKK872 pKa = 10.25RR873 pKa = 11.84RR874 pKa = 11.84VACCKK879 pKa = 10.03SEE881 pKa = 3.96PVARR885 pKa = 11.84LIMTLGHH892 pKa = 5.83VNYY895 pKa = 10.51KK896 pKa = 9.85HH897 pKa = 6.33VCEE900 pKa = 4.5CKK902 pKa = 10.24GDD904 pKa = 2.89IWRR907 pKa = 11.84CEE909 pKa = 3.85KK910 pKa = 10.9EE911 pKa = 4.25GVNGSGQCSTTSDD924 pKa = 3.28NTFSRR929 pKa = 11.84VGQARR934 pKa = 11.84ASGADD939 pKa = 3.12RR940 pKa = 11.84VIANGDD946 pKa = 3.47DD947 pKa = 3.27MAADD951 pKa = 4.26LGFDD955 pKa = 3.73PEE957 pKa = 3.99AAKK960 pKa = 10.94AFGTRR965 pKa = 11.84SRR967 pKa = 11.84DD968 pKa = 3.59VVVQDD973 pKa = 3.89ANLVPFTSHH982 pKa = 6.96HH983 pKa = 6.31INRR986 pKa = 11.84TTCTASYY993 pKa = 10.81DD994 pKa = 3.93RR995 pKa = 11.84PLKK998 pKa = 10.69LAFNLLSNCRR1008 pKa = 11.84SEE1010 pKa = 4.44DD1011 pKa = 2.95FGMRR1015 pKa = 11.84VDD1017 pKa = 3.21ALMYY1021 pKa = 9.49VVRR1024 pKa = 11.84NTPDD1028 pKa = 3.05ALAKK1032 pKa = 10.01FKK1034 pKa = 10.96KK1035 pKa = 9.97HH1036 pKa = 6.22LGPHH1040 pKa = 6.3AKK1042 pKa = 8.83GTDD1045 pKa = 3.58TCCVEE1050 pKa = 4.41MLWGAA1055 pKa = 3.62
Molecular weight: 116.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEB0|A0A1L3KEB0_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 3 OX=1922700 PE=4 SV=1
MM1 pKa = 7.2PRR3 pKa = 11.84RR4 pKa = 11.84WGRR7 pKa = 11.84SSPSDD12 pKa = 3.21AVLVTGVQQGAAAAVPKK29 pKa = 10.77AFGSTAGRR37 pKa = 11.84RR38 pKa = 11.84WGNRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84TSANSRR50 pKa = 11.84PNNRR54 pKa = 11.84WQRR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 9.66RR60 pKa = 11.84AGIAEE65 pKa = 4.39SACWDD70 pKa = 3.61AFSPVHH76 pKa = 5.97LALPRR81 pKa = 11.84AVAPYY86 pKa = 9.03ATIRR90 pKa = 11.84TTAIWNPDD98 pKa = 2.64VDD100 pKa = 4.86AGDD103 pKa = 3.86GNKK106 pKa = 9.56FALFGPYY113 pKa = 10.02LDD115 pKa = 4.68GSADD119 pKa = 3.32AGQWSNLFSLTANTALSNPKK139 pKa = 8.26NTNGGAKK146 pKa = 9.58YY147 pKa = 10.38YY148 pKa = 10.4AFEE151 pKa = 4.28SMISGSWKK159 pKa = 9.84AASVTPAAFSIQIMNPEE176 pKa = 4.17ALQTSSGAVYY186 pKa = 10.46IGRR189 pKa = 11.84CKK191 pKa = 10.75NRR193 pKa = 11.84VSVQEE198 pKa = 4.17GVLTEE203 pKa = 4.28TWQLLAEE210 pKa = 4.29EE211 pKa = 4.78LVSYY215 pKa = 10.19SNPRR219 pKa = 11.84ICSAGKK225 pKa = 9.08LALRR229 pKa = 11.84GVQIDD234 pKa = 4.76AIPNNMSEE242 pKa = 4.44LANFTTLKK250 pKa = 10.05EE251 pKa = 4.33YY252 pKa = 10.87SDD254 pKa = 3.6HH255 pKa = 5.66TTNLSSTVPHH265 pKa = 6.8PVHH268 pKa = 6.79PSGFNPIFVYY278 pKa = 10.85NPDD281 pKa = 3.5GVKK284 pKa = 10.47LQILVCCEE292 pKa = 3.01WRR294 pKa = 11.84VRR296 pKa = 11.84FDD298 pKa = 3.72PSNPAYY304 pKa = 10.09AACSSHH310 pKa = 6.88KK311 pKa = 9.14PTSDD315 pKa = 3.67TTWHH319 pKa = 6.14KK320 pKa = 10.59HH321 pKa = 4.15MQHH324 pKa = 6.5ALAAGNGVLDD334 pKa = 3.64IAEE337 pKa = 4.11KK338 pKa = 8.57VARR341 pKa = 11.84YY342 pKa = 8.16GVPIAKK348 pKa = 10.06AAGYY352 pKa = 10.32LGGAA356 pKa = 4.15
MM1 pKa = 7.2PRR3 pKa = 11.84RR4 pKa = 11.84WGRR7 pKa = 11.84SSPSDD12 pKa = 3.21AVLVTGVQQGAAAAVPKK29 pKa = 10.77AFGSTAGRR37 pKa = 11.84RR38 pKa = 11.84WGNRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84TSANSRR50 pKa = 11.84PNNRR54 pKa = 11.84WQRR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 9.66RR60 pKa = 11.84AGIAEE65 pKa = 4.39SACWDD70 pKa = 3.61AFSPVHH76 pKa = 5.97LALPRR81 pKa = 11.84AVAPYY86 pKa = 9.03ATIRR90 pKa = 11.84TTAIWNPDD98 pKa = 2.64VDD100 pKa = 4.86AGDD103 pKa = 3.86GNKK106 pKa = 9.56FALFGPYY113 pKa = 10.02LDD115 pKa = 4.68GSADD119 pKa = 3.32AGQWSNLFSLTANTALSNPKK139 pKa = 8.26NTNGGAKK146 pKa = 9.58YY147 pKa = 10.38YY148 pKa = 10.4AFEE151 pKa = 4.28SMISGSWKK159 pKa = 9.84AASVTPAAFSIQIMNPEE176 pKa = 4.17ALQTSSGAVYY186 pKa = 10.46IGRR189 pKa = 11.84CKK191 pKa = 10.75NRR193 pKa = 11.84VSVQEE198 pKa = 4.17GVLTEE203 pKa = 4.28TWQLLAEE210 pKa = 4.29EE211 pKa = 4.78LVSYY215 pKa = 10.19SNPRR219 pKa = 11.84ICSAGKK225 pKa = 9.08LALRR229 pKa = 11.84GVQIDD234 pKa = 4.76AIPNNMSEE242 pKa = 4.44LANFTTLKK250 pKa = 10.05EE251 pKa = 4.33YY252 pKa = 10.87SDD254 pKa = 3.6HH255 pKa = 5.66TTNLSSTVPHH265 pKa = 6.8PVHH268 pKa = 6.79PSGFNPIFVYY278 pKa = 10.85NPDD281 pKa = 3.5GVKK284 pKa = 10.47LQILVCCEE292 pKa = 3.01WRR294 pKa = 11.84VRR296 pKa = 11.84FDD298 pKa = 3.72PSNPAYY304 pKa = 10.09AACSSHH310 pKa = 6.88KK311 pKa = 9.14PTSDD315 pKa = 3.67TTWHH319 pKa = 6.14KK320 pKa = 10.59HH321 pKa = 4.15MQHH324 pKa = 6.5ALAAGNGVLDD334 pKa = 3.64IAEE337 pKa = 4.11KK338 pKa = 8.57VARR341 pKa = 11.84YY342 pKa = 8.16GVPIAKK348 pKa = 10.06AAGYY352 pKa = 10.32LGGAA356 pKa = 4.15
Molecular weight: 38.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1411 |
356 |
1055 |
705.5 |
77.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.993 ± 1.666 | 2.693 ± 0.481 |
6.378 ± 1.302 | 5.457 ± 1.13 |
4.04 ± 0.453 | 7.583 ± 0.135 |
2.197 ± 0.024 | 3.614 ± 0.152 |
5.386 ± 0.694 | 7.016 ± 0.265 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.268 ± 0.412 | 3.827 ± 1.123 |
4.819 ± 0.784 | 2.693 ± 0.189 |
5.953 ± 0.108 | 7.442 ± 0.739 |
5.67 ± 0.109 | 7.796 ± 0.637 |
1.701 ± 0.529 | 3.473 ± 0.183 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
