
Chryseolinea flava
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Fulvivirgaceae; Chryseolinea
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
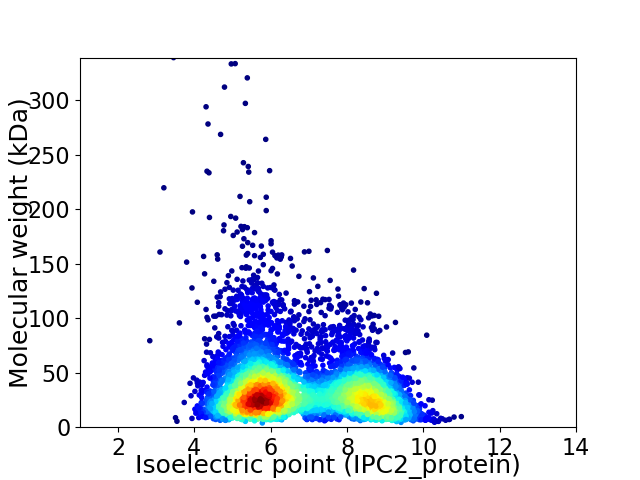
Virtual 2D-PAGE plot for 5446 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A364Y9H3|A0A364Y9H3_9BACT N-acetylglucosamine kinase OS=Chryseolinea flava OX=2059302 GN=DQQ10_02495 PE=4 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84FNLIYY7 pKa = 10.47SFAVAALFVCAFCAQPCFGQRR28 pKa = 11.84YY29 pKa = 7.87PLVFNSSSLDD39 pKa = 3.43GSNGFVMKK47 pKa = 10.67GNAANQVLGAEE58 pKa = 4.3LQFVGDD64 pKa = 3.99INNDD68 pKa = 3.35GLEE71 pKa = 4.35DD72 pKa = 3.76LAFSDD77 pKa = 3.99EE78 pKa = 4.17LEE80 pKa = 4.38EE81 pKa = 4.53VGGLALGGRR90 pKa = 11.84AYY92 pKa = 9.72IVFGSSTPFPKK103 pKa = 10.61EE104 pKa = 3.72FDD106 pKa = 3.57PKK108 pKa = 10.15TLNGSNGFMVDD119 pKa = 3.13PTTADD124 pKa = 3.1EE125 pKa = 5.09RR126 pKa = 11.84RR127 pKa = 11.84GQSIAGPGDD136 pKa = 3.43INGDD140 pKa = 3.97GIDD143 pKa = 4.78DD144 pKa = 4.84IIVSATSGEE153 pKa = 4.26LTMVIYY159 pKa = 10.35GRR161 pKa = 11.84SGAFPAVMKK170 pKa = 11.0VGDD173 pKa = 3.76INGTNGFVIQIDD185 pKa = 3.76GTTQVDD191 pKa = 3.51ALGDD195 pKa = 3.8MNGDD199 pKa = 4.55GINDD203 pKa = 3.85FAISNPHH210 pKa = 4.91WSEE213 pKa = 3.84RR214 pKa = 11.84AWVIFGRR221 pKa = 11.84SGNFPASINNTWLDD235 pKa = 3.64GVKK238 pKa = 10.88GFMTNPFPGSRR249 pKa = 11.84PAYY252 pKa = 9.22RR253 pKa = 11.84VGAAGDD259 pKa = 3.96INNDD263 pKa = 3.29GYY265 pKa = 11.65DD266 pKa = 3.82DD267 pKa = 4.84LLVGNWASASAEE279 pKa = 3.91AFSFLLFGRR288 pKa = 11.84ASFTSQVEE296 pKa = 4.31LTTLNGTDD304 pKa = 3.41GFRR307 pKa = 11.84IEE309 pKa = 4.18NAGNNFITCVGRR321 pKa = 11.84VGDD324 pKa = 3.99INGDD328 pKa = 3.51GMDD331 pKa = 4.41DD332 pKa = 3.94CFSEE336 pKa = 4.17NNLIFGSTSPFPSTLSMSSLNGNNGALLDD365 pKa = 4.54GYY367 pKa = 8.83VHH369 pKa = 6.87HH370 pKa = 7.29ACSAGDD376 pKa = 3.69LSGDD380 pKa = 4.1GIDD383 pKa = 4.39DD384 pKa = 4.39LLVVGQTGNYY394 pKa = 7.67VVFGTASGFPALVDD408 pKa = 3.84PEE410 pKa = 6.32DD411 pKa = 3.88IDD413 pKa = 3.79GTNGFTIPTSNTNIGRR429 pKa = 11.84PIDD432 pKa = 3.79GGKK435 pKa = 10.25DD436 pKa = 3.41FNGDD440 pKa = 3.38GFSDD444 pKa = 4.18FAYY447 pKa = 10.72DD448 pKa = 4.03NISGDD453 pKa = 3.39GTVAVVFGGDD463 pKa = 4.31HH464 pKa = 6.76IALPLNNNPSIIQLRR479 pKa = 11.84SDD481 pKa = 3.77GFRR484 pKa = 11.84IRR486 pKa = 11.84FNAKK490 pKa = 6.58EE491 pKa = 4.01TGIIRR496 pKa = 11.84YY497 pKa = 8.4AVYY500 pKa = 9.84PSSTPLMNNLDD511 pKa = 4.11VIDD514 pKa = 4.7NGTGAIVSGSFPIGSSNVDD533 pKa = 3.54IIHH536 pKa = 6.86LLDD539 pKa = 3.95GLAPDD544 pKa = 4.02TEE546 pKa = 4.56YY547 pKa = 11.37DD548 pKa = 3.91VYY550 pKa = 11.96VHH552 pKa = 6.92FLDD555 pKa = 5.09DD556 pKa = 5.44AGNKK560 pKa = 9.38DD561 pKa = 3.39INYY564 pKa = 8.64HH565 pKa = 5.49HH566 pKa = 7.66WDD568 pKa = 3.53NVKK571 pKa = 8.72TLPLGDD577 pKa = 3.78ITPPSITCPGNQTVVCTATTIPDD600 pKa = 3.27YY601 pKa = 10.12TALVTATDD609 pKa = 3.87NTDD612 pKa = 3.19PTPTVTQSPVSGSTFVNGMTITMTATDD639 pKa = 4.33DD640 pKa = 3.71AGNTQTCTFTVTKK653 pKa = 9.87TVDD656 pKa = 3.23AQKK659 pKa = 9.15PTITCPTDD667 pKa = 2.98RR668 pKa = 11.84TLSLNATIPDD678 pKa = 3.96YY679 pKa = 11.08RR680 pKa = 11.84SLATVTDD687 pKa = 3.64NCDD690 pKa = 3.05ASPTVTQSPVAGTIFTGPLTVTLTATDD717 pKa = 3.45ASSNVQTCSFFVGKK731 pKa = 10.37APDD734 pKa = 3.78TAPPTITCSGNQSLSCASTTIPSYY758 pKa = 11.33VSLATVSDD766 pKa = 4.23NEE768 pKa = 4.41DD769 pKa = 3.15ATPTVTQSPAAGSPFVDD786 pKa = 3.45GMTVTLTAIDD796 pKa = 4.13DD797 pKa = 4.12EE798 pKa = 5.49GNVASCNFIVAKK810 pKa = 8.37TTDD813 pKa = 3.42TQKK816 pKa = 11.05PIINCPGDD824 pKa = 3.55QVLTPGFVLPNYY836 pKa = 9.94VPTATTSDD844 pKa = 3.95NCDD847 pKa = 3.2MAPVVTQSPASGTMFITSQVVTLTVTDD874 pKa = 3.8ASGNSEE880 pKa = 3.83SCSFNVTPAPDD891 pKa = 3.63VEE893 pKa = 5.45DD894 pKa = 3.99PTISCPGNQSVSCTAITLPSYY915 pKa = 10.79ISLATVSDD923 pKa = 4.03NQDD926 pKa = 2.81ATPTVTQSPAAGSPVVTGMTVTLTATDD953 pKa = 3.99DD954 pKa = 3.92AGNTATCSFVVSKK967 pKa = 9.61TVDD970 pKa = 3.23AQKK973 pKa = 9.16PTITCPGNKK982 pKa = 9.23VLTIGEE988 pKa = 4.48TLPDD992 pKa = 3.79YY993 pKa = 10.81RR994 pKa = 11.84SLATAVDD1001 pKa = 3.84NCDD1004 pKa = 3.64LAPVITQSPASGGVFMAATVVTLTVTDD1031 pKa = 3.83ASGNSEE1037 pKa = 4.1TCSFTVTPAPDD1048 pKa = 3.52TEE1050 pKa = 4.97APAISCAGNQSVTCMTTSLPNYY1072 pKa = 9.06ISLATVSDD1080 pKa = 4.03NQDD1083 pKa = 2.99ATPTITQSPATGTPITNGMTVTLTATDD1110 pKa = 3.86DD1111 pKa = 3.82AGNSSTCSFMVAKK1124 pKa = 9.76PADD1127 pKa = 3.81TQKK1130 pKa = 11.12PIITCPANTTLNMGDD1145 pKa = 4.66AIADD1149 pKa = 3.72YY1150 pKa = 9.78TSQAIVADD1158 pKa = 4.04NCDD1161 pKa = 3.32ATPTVTQSPIAGTSFTGALTVTLTAKK1187 pKa = 10.26DD1188 pKa = 3.36ASGNTEE1194 pKa = 3.64VCSFSIAEE1202 pKa = 4.55APDD1205 pKa = 3.74SQSPTLTCPGDD1216 pKa = 3.49QLVDD1220 pKa = 3.7CDD1222 pKa = 3.8VTTVADD1228 pKa = 3.65YY1229 pKa = 9.85TSQVVASDD1237 pKa = 3.97NKK1239 pKa = 10.39DD1240 pKa = 3.11ASPTVTQSPAPGTTLTNGMTITLTAADD1267 pKa = 3.38IAGNEE1272 pKa = 4.44TTCSFTINVKK1282 pKa = 10.76SNVRR1286 pKa = 11.84VNAGRR1291 pKa = 11.84DD1292 pKa = 3.34VIMTVGGNVQLSAVASIDD1310 pKa = 3.15NGTFLWTPGEE1320 pKa = 4.11SLDD1323 pKa = 5.46RR1324 pKa = 11.84DD1325 pKa = 4.49DD1326 pKa = 5.22IPNPVATPLATTTYY1340 pKa = 10.03KK1341 pKa = 10.95VLFTTPNGCTATDD1354 pKa = 3.66EE1355 pKa = 4.65VKK1357 pKa = 10.98VEE1359 pKa = 4.2VKK1361 pKa = 10.34DD1362 pKa = 3.37LHH1364 pKa = 7.89IPEE1367 pKa = 4.96GFSPDD1372 pKa = 3.35GDD1374 pKa = 3.49GLNEE1378 pKa = 3.69NWILTGIEE1386 pKa = 4.01KK1387 pKa = 10.93YY1388 pKa = 8.82KK1389 pKa = 10.14TNEE1392 pKa = 3.47VNIFNRR1398 pKa = 11.84WGNCVYY1404 pKa = 10.56SVKK1407 pKa = 10.6GYY1409 pKa = 11.0NNQTLVFNGNANRR1422 pKa = 11.84LSALGADD1429 pKa = 4.0NLPDD1433 pKa = 3.24GTYY1436 pKa = 10.1FYY1438 pKa = 10.94SITLNDD1444 pKa = 3.21SSTPIKK1450 pKa = 10.74GFIVLKK1456 pKa = 10.59RR1457 pKa = 3.57
MM1 pKa = 7.96RR2 pKa = 11.84FNLIYY7 pKa = 10.47SFAVAALFVCAFCAQPCFGQRR28 pKa = 11.84YY29 pKa = 7.87PLVFNSSSLDD39 pKa = 3.43GSNGFVMKK47 pKa = 10.67GNAANQVLGAEE58 pKa = 4.3LQFVGDD64 pKa = 3.99INNDD68 pKa = 3.35GLEE71 pKa = 4.35DD72 pKa = 3.76LAFSDD77 pKa = 3.99EE78 pKa = 4.17LEE80 pKa = 4.38EE81 pKa = 4.53VGGLALGGRR90 pKa = 11.84AYY92 pKa = 9.72IVFGSSTPFPKK103 pKa = 10.61EE104 pKa = 3.72FDD106 pKa = 3.57PKK108 pKa = 10.15TLNGSNGFMVDD119 pKa = 3.13PTTADD124 pKa = 3.1EE125 pKa = 5.09RR126 pKa = 11.84RR127 pKa = 11.84GQSIAGPGDD136 pKa = 3.43INGDD140 pKa = 3.97GIDD143 pKa = 4.78DD144 pKa = 4.84IIVSATSGEE153 pKa = 4.26LTMVIYY159 pKa = 10.35GRR161 pKa = 11.84SGAFPAVMKK170 pKa = 11.0VGDD173 pKa = 3.76INGTNGFVIQIDD185 pKa = 3.76GTTQVDD191 pKa = 3.51ALGDD195 pKa = 3.8MNGDD199 pKa = 4.55GINDD203 pKa = 3.85FAISNPHH210 pKa = 4.91WSEE213 pKa = 3.84RR214 pKa = 11.84AWVIFGRR221 pKa = 11.84SGNFPASINNTWLDD235 pKa = 3.64GVKK238 pKa = 10.88GFMTNPFPGSRR249 pKa = 11.84PAYY252 pKa = 9.22RR253 pKa = 11.84VGAAGDD259 pKa = 3.96INNDD263 pKa = 3.29GYY265 pKa = 11.65DD266 pKa = 3.82DD267 pKa = 4.84LLVGNWASASAEE279 pKa = 3.91AFSFLLFGRR288 pKa = 11.84ASFTSQVEE296 pKa = 4.31LTTLNGTDD304 pKa = 3.41GFRR307 pKa = 11.84IEE309 pKa = 4.18NAGNNFITCVGRR321 pKa = 11.84VGDD324 pKa = 3.99INGDD328 pKa = 3.51GMDD331 pKa = 4.41DD332 pKa = 3.94CFSEE336 pKa = 4.17NNLIFGSTSPFPSTLSMSSLNGNNGALLDD365 pKa = 4.54GYY367 pKa = 8.83VHH369 pKa = 6.87HH370 pKa = 7.29ACSAGDD376 pKa = 3.69LSGDD380 pKa = 4.1GIDD383 pKa = 4.39DD384 pKa = 4.39LLVVGQTGNYY394 pKa = 7.67VVFGTASGFPALVDD408 pKa = 3.84PEE410 pKa = 6.32DD411 pKa = 3.88IDD413 pKa = 3.79GTNGFTIPTSNTNIGRR429 pKa = 11.84PIDD432 pKa = 3.79GGKK435 pKa = 10.25DD436 pKa = 3.41FNGDD440 pKa = 3.38GFSDD444 pKa = 4.18FAYY447 pKa = 10.72DD448 pKa = 4.03NISGDD453 pKa = 3.39GTVAVVFGGDD463 pKa = 4.31HH464 pKa = 6.76IALPLNNNPSIIQLRR479 pKa = 11.84SDD481 pKa = 3.77GFRR484 pKa = 11.84IRR486 pKa = 11.84FNAKK490 pKa = 6.58EE491 pKa = 4.01TGIIRR496 pKa = 11.84YY497 pKa = 8.4AVYY500 pKa = 9.84PSSTPLMNNLDD511 pKa = 4.11VIDD514 pKa = 4.7NGTGAIVSGSFPIGSSNVDD533 pKa = 3.54IIHH536 pKa = 6.86LLDD539 pKa = 3.95GLAPDD544 pKa = 4.02TEE546 pKa = 4.56YY547 pKa = 11.37DD548 pKa = 3.91VYY550 pKa = 11.96VHH552 pKa = 6.92FLDD555 pKa = 5.09DD556 pKa = 5.44AGNKK560 pKa = 9.38DD561 pKa = 3.39INYY564 pKa = 8.64HH565 pKa = 5.49HH566 pKa = 7.66WDD568 pKa = 3.53NVKK571 pKa = 8.72TLPLGDD577 pKa = 3.78ITPPSITCPGNQTVVCTATTIPDD600 pKa = 3.27YY601 pKa = 10.12TALVTATDD609 pKa = 3.87NTDD612 pKa = 3.19PTPTVTQSPVSGSTFVNGMTITMTATDD639 pKa = 4.33DD640 pKa = 3.71AGNTQTCTFTVTKK653 pKa = 9.87TVDD656 pKa = 3.23AQKK659 pKa = 9.15PTITCPTDD667 pKa = 2.98RR668 pKa = 11.84TLSLNATIPDD678 pKa = 3.96YY679 pKa = 11.08RR680 pKa = 11.84SLATVTDD687 pKa = 3.64NCDD690 pKa = 3.05ASPTVTQSPVAGTIFTGPLTVTLTATDD717 pKa = 3.45ASSNVQTCSFFVGKK731 pKa = 10.37APDD734 pKa = 3.78TAPPTITCSGNQSLSCASTTIPSYY758 pKa = 11.33VSLATVSDD766 pKa = 4.23NEE768 pKa = 4.41DD769 pKa = 3.15ATPTVTQSPAAGSPFVDD786 pKa = 3.45GMTVTLTAIDD796 pKa = 4.13DD797 pKa = 4.12EE798 pKa = 5.49GNVASCNFIVAKK810 pKa = 8.37TTDD813 pKa = 3.42TQKK816 pKa = 11.05PIINCPGDD824 pKa = 3.55QVLTPGFVLPNYY836 pKa = 9.94VPTATTSDD844 pKa = 3.95NCDD847 pKa = 3.2MAPVVTQSPASGTMFITSQVVTLTVTDD874 pKa = 3.8ASGNSEE880 pKa = 3.83SCSFNVTPAPDD891 pKa = 3.63VEE893 pKa = 5.45DD894 pKa = 3.99PTISCPGNQSVSCTAITLPSYY915 pKa = 10.79ISLATVSDD923 pKa = 4.03NQDD926 pKa = 2.81ATPTVTQSPAAGSPVVTGMTVTLTATDD953 pKa = 3.99DD954 pKa = 3.92AGNTATCSFVVSKK967 pKa = 9.61TVDD970 pKa = 3.23AQKK973 pKa = 9.16PTITCPGNKK982 pKa = 9.23VLTIGEE988 pKa = 4.48TLPDD992 pKa = 3.79YY993 pKa = 10.81RR994 pKa = 11.84SLATAVDD1001 pKa = 3.84NCDD1004 pKa = 3.64LAPVITQSPASGGVFMAATVVTLTVTDD1031 pKa = 3.83ASGNSEE1037 pKa = 4.1TCSFTVTPAPDD1048 pKa = 3.52TEE1050 pKa = 4.97APAISCAGNQSVTCMTTSLPNYY1072 pKa = 9.06ISLATVSDD1080 pKa = 4.03NQDD1083 pKa = 2.99ATPTITQSPATGTPITNGMTVTLTATDD1110 pKa = 3.86DD1111 pKa = 3.82AGNSSTCSFMVAKK1124 pKa = 9.76PADD1127 pKa = 3.81TQKK1130 pKa = 11.12PIITCPANTTLNMGDD1145 pKa = 4.66AIADD1149 pKa = 3.72YY1150 pKa = 9.78TSQAIVADD1158 pKa = 4.04NCDD1161 pKa = 3.32ATPTVTQSPIAGTSFTGALTVTLTAKK1187 pKa = 10.26DD1188 pKa = 3.36ASGNTEE1194 pKa = 3.64VCSFSIAEE1202 pKa = 4.55APDD1205 pKa = 3.74SQSPTLTCPGDD1216 pKa = 3.49QLVDD1220 pKa = 3.7CDD1222 pKa = 3.8VTTVADD1228 pKa = 3.65YY1229 pKa = 9.85TSQVVASDD1237 pKa = 3.97NKK1239 pKa = 10.39DD1240 pKa = 3.11ASPTVTQSPAPGTTLTNGMTITLTAADD1267 pKa = 3.38IAGNEE1272 pKa = 4.44TTCSFTINVKK1282 pKa = 10.76SNVRR1286 pKa = 11.84VNAGRR1291 pKa = 11.84DD1292 pKa = 3.34VIMTVGGNVQLSAVASIDD1310 pKa = 3.15NGTFLWTPGEE1320 pKa = 4.11SLDD1323 pKa = 5.46RR1324 pKa = 11.84DD1325 pKa = 4.49DD1326 pKa = 5.22IPNPVATPLATTTYY1340 pKa = 10.03KK1341 pKa = 10.95VLFTTPNGCTATDD1354 pKa = 3.66EE1355 pKa = 4.65VKK1357 pKa = 10.98VEE1359 pKa = 4.2VKK1361 pKa = 10.34DD1362 pKa = 3.37LHH1364 pKa = 7.89IPEE1367 pKa = 4.96GFSPDD1372 pKa = 3.35GDD1374 pKa = 3.49GLNEE1378 pKa = 3.69NWILTGIEE1386 pKa = 4.01KK1387 pKa = 10.93YY1388 pKa = 8.82KK1389 pKa = 10.14TNEE1392 pKa = 3.47VNIFNRR1398 pKa = 11.84WGNCVYY1404 pKa = 10.56SVKK1407 pKa = 10.6GYY1409 pKa = 11.0NNQTLVFNGNANRR1422 pKa = 11.84LSALGADD1429 pKa = 4.0NLPDD1433 pKa = 3.24GTYY1436 pKa = 10.1FYY1438 pKa = 10.94SITLNDD1444 pKa = 3.21SSTPIKK1450 pKa = 10.74GFIVLKK1456 pKa = 10.59RR1457 pKa = 3.57
Molecular weight: 151.43 kDa
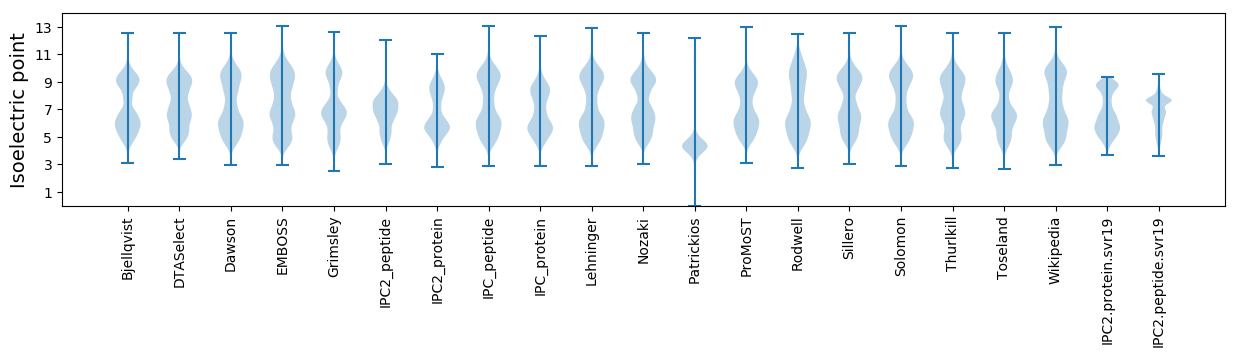
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A364XWY4|A0A364XWY4_9BACT Uncharacterized protein OS=Chryseolinea flava OX=2059302 GN=DQQ10_25885 PE=3 SV=1
MM1 pKa = 7.55AKK3 pKa = 10.01YY4 pKa = 9.99GKK6 pKa = 9.64KK7 pKa = 9.89AQSTVRR13 pKa = 11.84SAMKK17 pKa = 9.92RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.86KK21 pKa = 9.44GTLRR25 pKa = 11.84SGSGAKK31 pKa = 8.46VTSRR35 pKa = 11.84KK36 pKa = 8.12QAIAIGLSEE45 pKa = 4.44ARR47 pKa = 11.84QKK49 pKa = 9.87GAKK52 pKa = 9.43VPSKK56 pKa = 9.53KK57 pKa = 8.95TPSKK61 pKa = 9.79TRR63 pKa = 11.84AKK65 pKa = 10.15KK66 pKa = 9.07SARR69 pKa = 11.84RR70 pKa = 11.84KK71 pKa = 5.55PTKK74 pKa = 10.45RR75 pKa = 11.84MAKK78 pKa = 9.64RR79 pKa = 11.84SKK81 pKa = 8.96KK82 pKa = 6.56TARR85 pKa = 11.84AKK87 pKa = 10.44RR88 pKa = 3.58
MM1 pKa = 7.55AKK3 pKa = 10.01YY4 pKa = 9.99GKK6 pKa = 9.64KK7 pKa = 9.89AQSTVRR13 pKa = 11.84SAMKK17 pKa = 9.92RR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.86KK21 pKa = 9.44GTLRR25 pKa = 11.84SGSGAKK31 pKa = 8.46VTSRR35 pKa = 11.84KK36 pKa = 8.12QAIAIGLSEE45 pKa = 4.44ARR47 pKa = 11.84QKK49 pKa = 9.87GAKK52 pKa = 9.43VPSKK56 pKa = 9.53KK57 pKa = 8.95TPSKK61 pKa = 9.79TRR63 pKa = 11.84AKK65 pKa = 10.15KK66 pKa = 9.07SARR69 pKa = 11.84RR70 pKa = 11.84KK71 pKa = 5.55PTKK74 pKa = 10.45RR75 pKa = 11.84MAKK78 pKa = 9.64RR79 pKa = 11.84SKK81 pKa = 8.96KK82 pKa = 6.56TARR85 pKa = 11.84AKK87 pKa = 10.44RR88 pKa = 3.58
Molecular weight: 9.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1947148 |
33 |
3382 |
357.5 |
40.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.415 ± 0.033 | 0.759 ± 0.012 |
5.576 ± 0.025 | 5.835 ± 0.036 |
5.058 ± 0.025 | 6.738 ± 0.035 |
2.034 ± 0.017 | 7.04 ± 0.024 |
6.618 ± 0.04 | 9.14 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.288 ± 0.015 | 5.219 ± 0.032 |
3.71 ± 0.023 | 3.637 ± 0.019 |
4.352 ± 0.023 | 6.563 ± 0.029 |
6.058 ± 0.045 | 6.855 ± 0.025 |
1.234 ± 0.012 | 3.873 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
