
Halorhodospira halochloris (Ectothiorhodospira halochloris)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Halorhodospira
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
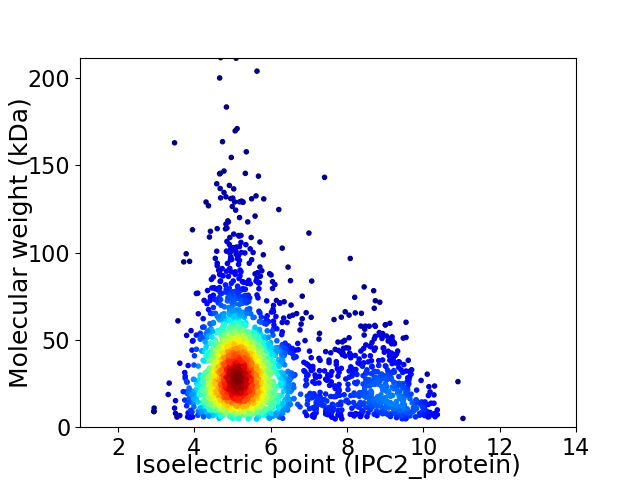
Virtual 2D-PAGE plot for 2566 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0X8XA14|A0A0X8XA14_HALHR Uncharacterized protein OS=Halorhodospira halochloris OX=1052 GN=HH1059_15450 PE=4 SV=1
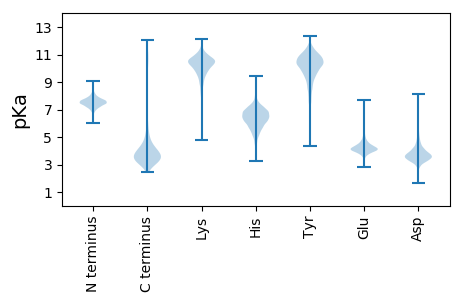
MM1 pKa = 6.98PQLRR5 pKa = 11.84RR6 pKa = 11.84LYY8 pKa = 9.13PAAIALSSAVCTGSLSAAEE27 pKa = 4.56RR28 pKa = 11.84FTGGYY33 pKa = 9.36IGVEE37 pKa = 3.89GDD39 pKa = 3.85LLSSVTFEE47 pKa = 4.95RR48 pKa = 11.84DD49 pKa = 2.92FDD51 pKa = 4.33DD52 pKa = 5.04GFAGEE57 pKa = 4.29VVEE60 pKa = 6.21DD61 pKa = 4.01FVEE64 pKa = 4.7YY65 pKa = 10.6EE66 pKa = 4.31DD67 pKa = 4.04PTDD70 pKa = 3.9FLEE73 pKa = 6.08NDD75 pKa = 3.61DD76 pKa = 3.93QQEE79 pKa = 4.37GFASALMYY87 pKa = 9.75TSHH90 pKa = 6.78EE91 pKa = 4.13AGIEE95 pKa = 4.03EE96 pKa = 4.55VVPGGSVILGGGMQQDD112 pKa = 3.79GFYY115 pKa = 10.68SGIEE119 pKa = 3.77ARR121 pKa = 11.84YY122 pKa = 8.98HH123 pKa = 5.79FGGVDD128 pKa = 3.36EE129 pKa = 4.95EE130 pKa = 5.13FEE132 pKa = 4.97DD133 pKa = 4.8DD134 pKa = 4.67LEE136 pKa = 4.41EE137 pKa = 4.45SLEE140 pKa = 4.13FEE142 pKa = 4.78DD143 pKa = 6.07GYY145 pKa = 11.13SISARR150 pKa = 11.84FGTLLRR156 pKa = 11.84EE157 pKa = 4.5GNVMLYY163 pKa = 10.95GSLGYY168 pKa = 8.81ATRR171 pKa = 11.84EE172 pKa = 3.56VTYY175 pKa = 10.51EE176 pKa = 3.93VFEE179 pKa = 5.45DD180 pKa = 5.25DD181 pKa = 5.14EE182 pKa = 5.66NSDD185 pKa = 4.26SNDD188 pKa = 2.93HH189 pKa = 6.17SGFRR193 pKa = 11.84AGIGLEE199 pKa = 3.95YY200 pKa = 10.59RR201 pKa = 11.84PDD203 pKa = 3.48YY204 pKa = 10.83PPIFVRR210 pKa = 11.84LEE212 pKa = 3.74TSRR215 pKa = 11.84TDD217 pKa = 4.53FGDD220 pKa = 3.25EE221 pKa = 4.22TYY223 pKa = 11.22EE224 pKa = 5.6DD225 pKa = 4.18EE226 pKa = 6.03DD227 pKa = 5.25DD228 pKa = 4.52GNEE231 pKa = 4.31FDD233 pKa = 6.02FADD236 pKa = 3.54IDD238 pKa = 4.41DD239 pKa = 4.61LVEE242 pKa = 4.16HH243 pKa = 6.97AAHH246 pKa = 6.77LGVGYY251 pKa = 10.61RR252 pKa = 11.84FF253 pKa = 3.62
MM1 pKa = 6.98PQLRR5 pKa = 11.84RR6 pKa = 11.84LYY8 pKa = 9.13PAAIALSSAVCTGSLSAAEE27 pKa = 4.56RR28 pKa = 11.84FTGGYY33 pKa = 9.36IGVEE37 pKa = 3.89GDD39 pKa = 3.85LLSSVTFEE47 pKa = 4.95RR48 pKa = 11.84DD49 pKa = 2.92FDD51 pKa = 4.33DD52 pKa = 5.04GFAGEE57 pKa = 4.29VVEE60 pKa = 6.21DD61 pKa = 4.01FVEE64 pKa = 4.7YY65 pKa = 10.6EE66 pKa = 4.31DD67 pKa = 4.04PTDD70 pKa = 3.9FLEE73 pKa = 6.08NDD75 pKa = 3.61DD76 pKa = 3.93QQEE79 pKa = 4.37GFASALMYY87 pKa = 9.75TSHH90 pKa = 6.78EE91 pKa = 4.13AGIEE95 pKa = 4.03EE96 pKa = 4.55VVPGGSVILGGGMQQDD112 pKa = 3.79GFYY115 pKa = 10.68SGIEE119 pKa = 3.77ARR121 pKa = 11.84YY122 pKa = 8.98HH123 pKa = 5.79FGGVDD128 pKa = 3.36EE129 pKa = 4.95EE130 pKa = 5.13FEE132 pKa = 4.97DD133 pKa = 4.8DD134 pKa = 4.67LEE136 pKa = 4.41EE137 pKa = 4.45SLEE140 pKa = 4.13FEE142 pKa = 4.78DD143 pKa = 6.07GYY145 pKa = 11.13SISARR150 pKa = 11.84FGTLLRR156 pKa = 11.84EE157 pKa = 4.5GNVMLYY163 pKa = 10.95GSLGYY168 pKa = 8.81ATRR171 pKa = 11.84EE172 pKa = 3.56VTYY175 pKa = 10.51EE176 pKa = 3.93VFEE179 pKa = 5.45DD180 pKa = 5.25DD181 pKa = 5.14EE182 pKa = 5.66NSDD185 pKa = 4.26SNDD188 pKa = 2.93HH189 pKa = 6.17SGFRR193 pKa = 11.84AGIGLEE199 pKa = 3.95YY200 pKa = 10.59RR201 pKa = 11.84PDD203 pKa = 3.48YY204 pKa = 10.83PPIFVRR210 pKa = 11.84LEE212 pKa = 3.74TSRR215 pKa = 11.84TDD217 pKa = 4.53FGDD220 pKa = 3.25EE221 pKa = 4.22TYY223 pKa = 11.22EE224 pKa = 5.6DD225 pKa = 4.18EE226 pKa = 6.03DD227 pKa = 5.25DD228 pKa = 4.52GNEE231 pKa = 4.31FDD233 pKa = 6.02FADD236 pKa = 3.54IDD238 pKa = 4.41DD239 pKa = 4.61LVEE242 pKa = 4.16HH243 pKa = 6.97AAHH246 pKa = 6.77LGVGYY251 pKa = 10.61RR252 pKa = 11.84FF253 pKa = 3.62
Molecular weight: 28.04 kDa
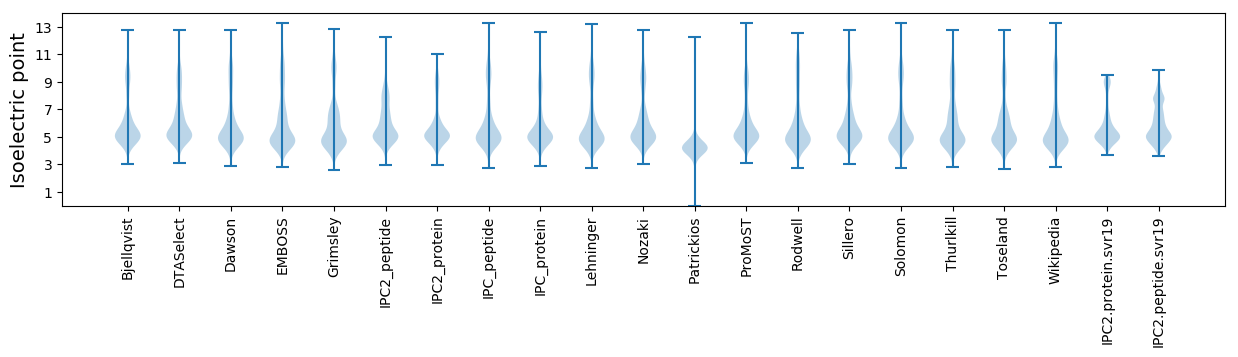
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6EZW3|A0A2Z6EZW3_HALHR Uncharacterized protein OS=Halorhodospira halochloris OX=1052 GN=HH1059_19610 PE=4 SV=1
MM1 pKa = 7.72AKK3 pKa = 10.09PRR5 pKa = 11.84GNRR8 pKa = 11.84VTVTRR13 pKa = 11.84RR14 pKa = 11.84TSKK17 pKa = 9.53RR18 pKa = 11.84TQALVKK24 pKa = 10.06APHH27 pKa = 6.94ILRR30 pKa = 11.84SNKK33 pKa = 9.43GNSPLKK39 pKa = 10.01VIRR42 pKa = 11.84VALL45 pKa = 3.92
MM1 pKa = 7.72AKK3 pKa = 10.09PRR5 pKa = 11.84GNRR8 pKa = 11.84VTVTRR13 pKa = 11.84RR14 pKa = 11.84TSKK17 pKa = 9.53RR18 pKa = 11.84TQALVKK24 pKa = 10.06APHH27 pKa = 6.94ILRR30 pKa = 11.84SNKK33 pKa = 9.43GNSPLKK39 pKa = 10.01VIRR42 pKa = 11.84VALL45 pKa = 3.92
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
818812 |
43 |
1910 |
319.1 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.48 ± 0.065 | 0.97 ± 0.015 |
6.015 ± 0.046 | 7.571 ± 0.054 |
3.232 ± 0.026 | 8.016 ± 0.045 |
2.402 ± 0.025 | 5.124 ± 0.035 |
2.861 ± 0.034 | 10.473 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.142 ± 0.023 | 2.776 ± 0.03 |
4.707 ± 0.033 | 4.429 ± 0.035 |
7.498 ± 0.052 | 5.804 ± 0.032 |
4.591 ± 0.03 | 6.903 ± 0.045 |
1.378 ± 0.02 | 2.629 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
