
Escherichia phage K1ind1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Guernseyvirinae; Kagunavirus; Escherichia virus K1ind1
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
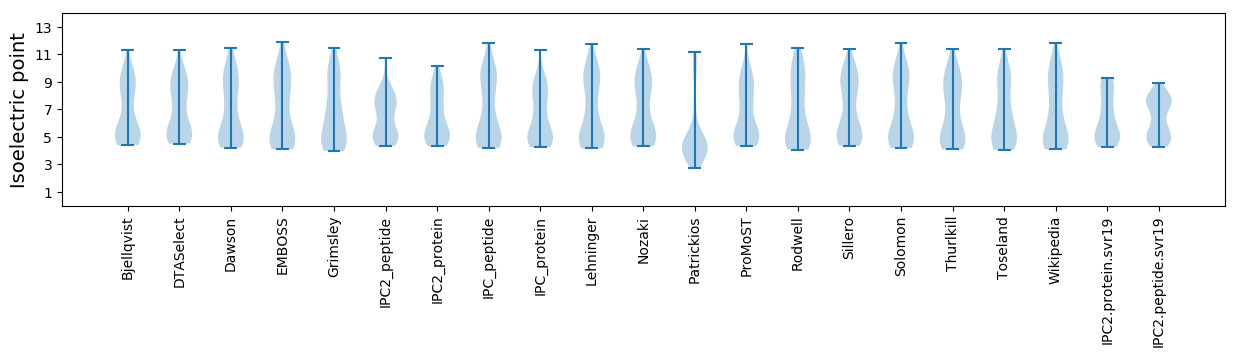
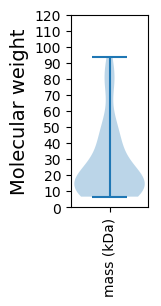
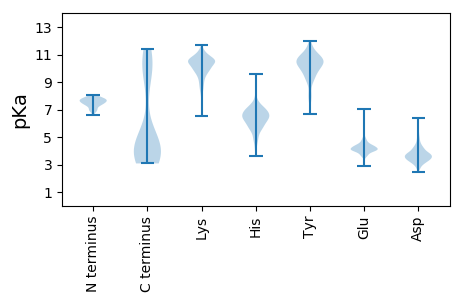
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2XJJ3|D2XJJ3_9CAUD Uncharacterized protein OS=Escherichia phage K1ind1 OX=698488 PE=4 SV=1
MM1 pKa = 7.76SDD3 pKa = 3.72LYY5 pKa = 10.8FPRR8 pKa = 11.84SLKK11 pKa = 10.58PIVSKK16 pKa = 10.6GYY18 pKa = 11.11SMTRR22 pKa = 11.84RR23 pKa = 11.84NNVWSVDD30 pKa = 3.73LAGGGVRR37 pKa = 11.84QGRR40 pKa = 11.84DD41 pKa = 2.51TYY43 pKa = 11.45YY44 pKa = 11.05DD45 pKa = 3.59VFPVSVTLITSAMGRR60 pKa = 11.84QAFLSFLEE68 pKa = 4.46KK69 pKa = 10.56VDD71 pKa = 4.19GGASSFWMAHH81 pKa = 5.89DD82 pKa = 4.3FGMGIEE88 pKa = 4.86DD89 pKa = 3.88YY90 pKa = 10.84QVTITSTIAEE100 pKa = 4.27STEE103 pKa = 4.87DD104 pKa = 4.05GINWTITFTATAEE117 pKa = 4.04KK118 pKa = 10.95SPFQDD123 pKa = 5.19LEE125 pKa = 4.27NQCLINNLPDD135 pKa = 4.42LYY137 pKa = 11.04GCYY140 pKa = 10.16GDD142 pKa = 4.96CLGSFLKK149 pKa = 10.31IYY151 pKa = 10.89ANYY154 pKa = 7.19EE155 pKa = 4.02TTFPRR160 pKa = 11.84IWSNEE165 pKa = 3.52GPAGYY170 pKa = 9.29PPINLLASTLDD181 pKa = 3.34SRR183 pKa = 11.84VVYY186 pKa = 10.23DD187 pKa = 4.5GPQVYY192 pKa = 10.4YY193 pKa = 10.37INRR196 pKa = 11.84NGNLVQSAANEE207 pKa = 4.02WPLTFIDD214 pKa = 4.87GSAVGRR220 pKa = 11.84VPPEE224 pKa = 3.51VSVTNRR230 pKa = 11.84FLYY233 pKa = 10.64SSNFSSSNWNKK244 pKa = 10.0GLVSVSVSEE253 pKa = 4.29TGVLTAAGTYY263 pKa = 10.49DD264 pKa = 3.4VTATTTSGNHH274 pKa = 7.46LINQTVDD281 pKa = 3.07VSVGVGDD288 pKa = 3.66MVTFSFFAKK297 pKa = 10.21ANGYY301 pKa = 9.34NFIRR305 pKa = 11.84MYY307 pKa = 10.64VRR309 pKa = 11.84EE310 pKa = 4.67LPTVLGSAVVDD321 pKa = 4.12LRR323 pKa = 11.84DD324 pKa = 3.31GSIYY328 pKa = 10.47SGPGGTDD335 pKa = 3.01VTDD338 pKa = 3.58VGGGWFRR345 pKa = 11.84ISEE348 pKa = 4.6TITATEE354 pKa = 4.63DD355 pKa = 2.89KK356 pKa = 10.49TMSIRR361 pKa = 11.84MDD363 pKa = 2.59AWIYY367 pKa = 10.54SDD369 pKa = 3.93GVTGSFAGDD378 pKa = 3.37GVSGIKK384 pKa = 10.44LSAATVEE391 pKa = 4.51VNQNTTSPVITDD403 pKa = 3.35AVVATRR409 pKa = 11.84TTASAKK415 pKa = 8.86VTMNGATSIDD425 pKa = 3.23IAYY428 pKa = 10.57SDD430 pKa = 3.69GSVVNVPSVDD440 pKa = 3.72GYY442 pKa = 11.97ASIPQADD449 pKa = 4.13SAWGSKK455 pKa = 10.25YY456 pKa = 8.24ITRR459 pKa = 11.84IDD461 pKa = 3.86FNVDD465 pKa = 2.67GG466 pKa = 4.77
MM1 pKa = 7.76SDD3 pKa = 3.72LYY5 pKa = 10.8FPRR8 pKa = 11.84SLKK11 pKa = 10.58PIVSKK16 pKa = 10.6GYY18 pKa = 11.11SMTRR22 pKa = 11.84RR23 pKa = 11.84NNVWSVDD30 pKa = 3.73LAGGGVRR37 pKa = 11.84QGRR40 pKa = 11.84DD41 pKa = 2.51TYY43 pKa = 11.45YY44 pKa = 11.05DD45 pKa = 3.59VFPVSVTLITSAMGRR60 pKa = 11.84QAFLSFLEE68 pKa = 4.46KK69 pKa = 10.56VDD71 pKa = 4.19GGASSFWMAHH81 pKa = 5.89DD82 pKa = 4.3FGMGIEE88 pKa = 4.86DD89 pKa = 3.88YY90 pKa = 10.84QVTITSTIAEE100 pKa = 4.27STEE103 pKa = 4.87DD104 pKa = 4.05GINWTITFTATAEE117 pKa = 4.04KK118 pKa = 10.95SPFQDD123 pKa = 5.19LEE125 pKa = 4.27NQCLINNLPDD135 pKa = 4.42LYY137 pKa = 11.04GCYY140 pKa = 10.16GDD142 pKa = 4.96CLGSFLKK149 pKa = 10.31IYY151 pKa = 10.89ANYY154 pKa = 7.19EE155 pKa = 4.02TTFPRR160 pKa = 11.84IWSNEE165 pKa = 3.52GPAGYY170 pKa = 9.29PPINLLASTLDD181 pKa = 3.34SRR183 pKa = 11.84VVYY186 pKa = 10.23DD187 pKa = 4.5GPQVYY192 pKa = 10.4YY193 pKa = 10.37INRR196 pKa = 11.84NGNLVQSAANEE207 pKa = 4.02WPLTFIDD214 pKa = 4.87GSAVGRR220 pKa = 11.84VPPEE224 pKa = 3.51VSVTNRR230 pKa = 11.84FLYY233 pKa = 10.64SSNFSSSNWNKK244 pKa = 10.0GLVSVSVSEE253 pKa = 4.29TGVLTAAGTYY263 pKa = 10.49DD264 pKa = 3.4VTATTTSGNHH274 pKa = 7.46LINQTVDD281 pKa = 3.07VSVGVGDD288 pKa = 3.66MVTFSFFAKK297 pKa = 10.21ANGYY301 pKa = 9.34NFIRR305 pKa = 11.84MYY307 pKa = 10.64VRR309 pKa = 11.84EE310 pKa = 4.67LPTVLGSAVVDD321 pKa = 4.12LRR323 pKa = 11.84DD324 pKa = 3.31GSIYY328 pKa = 10.47SGPGGTDD335 pKa = 3.01VTDD338 pKa = 3.58VGGGWFRR345 pKa = 11.84ISEE348 pKa = 4.6TITATEE354 pKa = 4.63DD355 pKa = 2.89KK356 pKa = 10.49TMSIRR361 pKa = 11.84MDD363 pKa = 2.59AWIYY367 pKa = 10.54SDD369 pKa = 3.93GVTGSFAGDD378 pKa = 3.37GVSGIKK384 pKa = 10.44LSAATVEE391 pKa = 4.51VNQNTTSPVITDD403 pKa = 3.35AVVATRR409 pKa = 11.84TTASAKK415 pKa = 8.86VTMNGATSIDD425 pKa = 3.23IAYY428 pKa = 10.57SDD430 pKa = 3.69GSVVNVPSVDD440 pKa = 3.72GYY442 pKa = 11.97ASIPQADD449 pKa = 4.13SAWGSKK455 pKa = 10.25YY456 pKa = 8.24ITRR459 pKa = 11.84IDD461 pKa = 3.86FNVDD465 pKa = 2.67GG466 pKa = 4.77
Molecular weight: 50.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2XJK0|D2XJK0_9CAUD Putative DNA polymerase OS=Escherichia phage K1ind1 OX=698488 PE=3 SV=1
MM1 pKa = 7.77KK2 pKa = 9.71EE3 pKa = 4.14ALPKK7 pKa = 10.57LSQKK11 pKa = 9.0TLTGSPSVIFSRR23 pKa = 11.84VSAAGLSRR31 pKa = 11.84SDD33 pKa = 3.17LRR35 pKa = 11.84EE36 pKa = 3.59FRR38 pKa = 11.84KK39 pKa = 9.09MRR41 pKa = 11.84LSGRR45 pKa = 11.84EE46 pKa = 3.5ARR48 pKa = 11.84LANRR52 pKa = 11.84LARR55 pKa = 11.84RR56 pKa = 11.84EE57 pKa = 4.31SNSDD61 pKa = 2.97SSTLDD66 pKa = 3.31TLHH69 pKa = 6.73LCSSTSSASAALQSSLGNRR88 pKa = 11.84LRR90 pKa = 11.84QQLQNRR96 pKa = 11.84GCAIYY101 pKa = 10.63RR102 pKa = 11.84FTWKK106 pKa = 10.52EE107 pKa = 3.48KK108 pKa = 6.65TTPSGLPYY116 pKa = 9.93FQLVASAHH124 pKa = 4.83RR125 pKa = 11.84TKK127 pKa = 10.84EE128 pKa = 4.1SGSSLVRR135 pKa = 11.84TNWPTPTANDD145 pKa = 3.38YY146 pKa = 10.56RR147 pKa = 11.84GSGEE151 pKa = 4.02TVIRR155 pKa = 11.84KK156 pKa = 9.32DD157 pKa = 3.34GKK159 pKa = 10.49DD160 pKa = 3.04RR161 pKa = 11.84TFDD164 pKa = 3.46RR165 pKa = 11.84LDD167 pKa = 3.27YY168 pKa = 10.57STEE171 pKa = 3.76QGLKK175 pKa = 7.4ITQPIRR181 pKa = 11.84ITASGQILTGSDD193 pKa = 2.85AGMEE197 pKa = 4.07NSGQLNPAHH206 pKa = 6.46SRR208 pKa = 11.84WLMGYY213 pKa = 8.97PPEE216 pKa = 4.38WDD218 pKa = 3.7DD219 pKa = 4.05CAVMAMPSSRR229 pKa = 11.84KK230 pKa = 9.53
MM1 pKa = 7.77KK2 pKa = 9.71EE3 pKa = 4.14ALPKK7 pKa = 10.57LSQKK11 pKa = 9.0TLTGSPSVIFSRR23 pKa = 11.84VSAAGLSRR31 pKa = 11.84SDD33 pKa = 3.17LRR35 pKa = 11.84EE36 pKa = 3.59FRR38 pKa = 11.84KK39 pKa = 9.09MRR41 pKa = 11.84LSGRR45 pKa = 11.84EE46 pKa = 3.5ARR48 pKa = 11.84LANRR52 pKa = 11.84LARR55 pKa = 11.84RR56 pKa = 11.84EE57 pKa = 4.31SNSDD61 pKa = 2.97SSTLDD66 pKa = 3.31TLHH69 pKa = 6.73LCSSTSSASAALQSSLGNRR88 pKa = 11.84LRR90 pKa = 11.84QQLQNRR96 pKa = 11.84GCAIYY101 pKa = 10.63RR102 pKa = 11.84FTWKK106 pKa = 10.52EE107 pKa = 3.48KK108 pKa = 6.65TTPSGLPYY116 pKa = 9.93FQLVASAHH124 pKa = 4.83RR125 pKa = 11.84TKK127 pKa = 10.84EE128 pKa = 4.1SGSSLVRR135 pKa = 11.84TNWPTPTANDD145 pKa = 3.38YY146 pKa = 10.56RR147 pKa = 11.84GSGEE151 pKa = 4.02TVIRR155 pKa = 11.84KK156 pKa = 9.32DD157 pKa = 3.34GKK159 pKa = 10.49DD160 pKa = 3.04RR161 pKa = 11.84TFDD164 pKa = 3.46RR165 pKa = 11.84LDD167 pKa = 3.27YY168 pKa = 10.57STEE171 pKa = 3.76QGLKK175 pKa = 7.4ITQPIRR181 pKa = 11.84ITASGQILTGSDD193 pKa = 2.85AGMEE197 pKa = 4.07NSGQLNPAHH206 pKa = 6.46SRR208 pKa = 11.84WLMGYY213 pKa = 8.97PPEE216 pKa = 4.38WDD218 pKa = 3.7DD219 pKa = 4.05CAVMAMPSSRR229 pKa = 11.84KK230 pKa = 9.53
Molecular weight: 25.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12555 |
56 |
851 |
246.2 |
27.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.98 ± 0.501 | 1.091 ± 0.142 |
6.197 ± 0.263 | 6.34 ± 0.494 |
3.863 ± 0.178 | 7.869 ± 0.291 |
1.728 ± 0.195 | 5.217 ± 0.152 |
5.504 ± 0.393 | 7.806 ± 0.278 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.374 ± 0.174 | 4.19 ± 0.26 |
3.903 ± 0.251 | 3.775 ± 0.351 |
5.368 ± 0.281 | 6.476 ± 0.518 |
6.571 ± 0.389 | 7.121 ± 0.333 |
1.33 ± 0.159 | 3.297 ± 0.222 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
