
Bacillus hemicellulosilyticus JCM 9152
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalihalobacillus; Alkalihalobacillus hemicellulosilyticus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
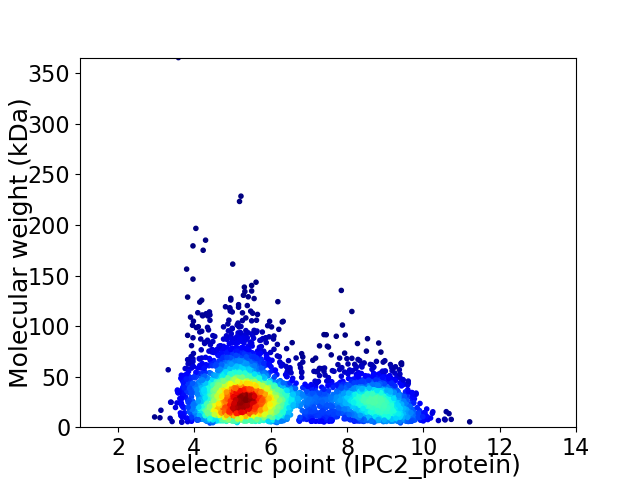
Virtual 2D-PAGE plot for 4345 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4QEQ1|W4QEQ1_9BACI rRNA methylase OS=Bacillus hemicellulosilyticus JCM 9152 OX=1236971 GN=JCM9152_1545 PE=4 SV=1
MM1 pKa = 6.95MKK3 pKa = 10.07KK4 pKa = 10.19GFVFIVLLTFMLVIVACNNSEE25 pKa = 4.2VTGEE29 pKa = 4.21EE30 pKa = 4.67GNDD33 pKa = 3.43SDD35 pKa = 6.04DD36 pKa = 3.63STNEE40 pKa = 3.82VEE42 pKa = 6.35NADD45 pKa = 3.59QDD47 pKa = 3.77QAADD51 pKa = 4.48DD52 pKa = 4.13EE53 pKa = 4.68AQNTDD58 pKa = 3.01GPVTVTFWHH67 pKa = 6.43AMGGALQEE75 pKa = 4.5ALDD78 pKa = 4.4EE79 pKa = 4.22LVNMYY84 pKa = 10.2NAAQDD89 pKa = 3.84DD90 pKa = 4.82VIVEE94 pKa = 4.16AEE96 pKa = 4.12YY97 pKa = 10.67QGSYY101 pKa = 11.29DD102 pKa = 3.46EE103 pKa = 5.51ALTKK107 pKa = 9.98FHH109 pKa = 6.95SVAGTDD115 pKa = 3.53SAPTMIQVFEE125 pKa = 4.42IGTMSMVHH133 pKa = 6.33SGHH136 pKa = 6.38ITPIQDD142 pKa = 4.88LIDD145 pKa = 4.26ADD147 pKa = 4.55GYY149 pKa = 11.67DD150 pKa = 3.65MSSLEE155 pKa = 4.35DD156 pKa = 3.85NIINYY161 pKa = 9.14YY162 pKa = 10.46KK163 pKa = 10.06IDD165 pKa = 3.66DD166 pKa = 4.03QFYY169 pKa = 10.74SMPFNSSTPVMYY181 pKa = 10.65YY182 pKa = 10.75NKK184 pKa = 10.28DD185 pKa = 3.16AFEE188 pKa = 4.36AAGLDD193 pKa = 3.61PDD195 pKa = 4.94APPEE199 pKa = 4.03TYY201 pKa = 10.61EE202 pKa = 4.32EE203 pKa = 4.15IEE205 pKa = 4.01EE206 pKa = 4.06ASRR209 pKa = 11.84QIVEE213 pKa = 4.9SNPDD217 pKa = 3.17MKK219 pKa = 11.03GFALQAYY226 pKa = 7.7GWLYY230 pKa = 10.57EE231 pKa = 4.13QLLANQGALLMNNDD245 pKa = 3.55NGRR248 pKa = 11.84NGTPTEE254 pKa = 4.09VGFTEE259 pKa = 4.96AEE261 pKa = 4.35GKK263 pKa = 10.06SVFAWVEE270 pKa = 3.66RR271 pKa = 11.84MIEE274 pKa = 4.13DD275 pKa = 3.88DD276 pKa = 3.41TFANYY281 pKa = 7.44GTNSDD286 pKa = 3.57NMVSGFMSGDD296 pKa = 2.87VAMFLQSSASVKK308 pKa = 10.49NVVEE312 pKa = 3.97NAPFEE317 pKa = 4.42VGVAFLPYY325 pKa = 10.08PEE327 pKa = 3.93NTEE330 pKa = 3.96RR331 pKa = 11.84NGVVIGGASLWMVDD345 pKa = 3.69GKK347 pKa = 10.26PEE349 pKa = 4.18AEE351 pKa = 5.5QMAAWDD357 pKa = 4.16FMKK360 pKa = 10.5FLQTPEE366 pKa = 4.39VQAQWHH372 pKa = 5.97VGTGYY377 pKa = 10.49FAINPAAYY385 pKa = 9.91DD386 pKa = 3.67EE387 pKa = 4.46EE388 pKa = 4.9VVLEE392 pKa = 4.67AYY394 pKa = 7.67EE395 pKa = 4.14TMPQLKK401 pKa = 8.25VTVDD405 pKa = 3.2QLQATTSNYY414 pKa = 7.76ATQGAIMDD422 pKa = 5.0MIPEE426 pKa = 4.09ARR428 pKa = 11.84RR429 pKa = 11.84ITEE432 pKa = 3.98TALEE436 pKa = 4.18TVYY439 pKa = 11.26NGGAVDD445 pKa = 3.98DD446 pKa = 4.9TYY448 pKa = 11.73QSVVDD453 pKa = 4.52QINAAIEE460 pKa = 3.89QANRR464 pKa = 11.84ARR466 pKa = 11.84GEE468 pKa = 3.89
MM1 pKa = 6.95MKK3 pKa = 10.07KK4 pKa = 10.19GFVFIVLLTFMLVIVACNNSEE25 pKa = 4.2VTGEE29 pKa = 4.21EE30 pKa = 4.67GNDD33 pKa = 3.43SDD35 pKa = 6.04DD36 pKa = 3.63STNEE40 pKa = 3.82VEE42 pKa = 6.35NADD45 pKa = 3.59QDD47 pKa = 3.77QAADD51 pKa = 4.48DD52 pKa = 4.13EE53 pKa = 4.68AQNTDD58 pKa = 3.01GPVTVTFWHH67 pKa = 6.43AMGGALQEE75 pKa = 4.5ALDD78 pKa = 4.4EE79 pKa = 4.22LVNMYY84 pKa = 10.2NAAQDD89 pKa = 3.84DD90 pKa = 4.82VIVEE94 pKa = 4.16AEE96 pKa = 4.12YY97 pKa = 10.67QGSYY101 pKa = 11.29DD102 pKa = 3.46EE103 pKa = 5.51ALTKK107 pKa = 9.98FHH109 pKa = 6.95SVAGTDD115 pKa = 3.53SAPTMIQVFEE125 pKa = 4.42IGTMSMVHH133 pKa = 6.33SGHH136 pKa = 6.38ITPIQDD142 pKa = 4.88LIDD145 pKa = 4.26ADD147 pKa = 4.55GYY149 pKa = 11.67DD150 pKa = 3.65MSSLEE155 pKa = 4.35DD156 pKa = 3.85NIINYY161 pKa = 9.14YY162 pKa = 10.46KK163 pKa = 10.06IDD165 pKa = 3.66DD166 pKa = 4.03QFYY169 pKa = 10.74SMPFNSSTPVMYY181 pKa = 10.65YY182 pKa = 10.75NKK184 pKa = 10.28DD185 pKa = 3.16AFEE188 pKa = 4.36AAGLDD193 pKa = 3.61PDD195 pKa = 4.94APPEE199 pKa = 4.03TYY201 pKa = 10.61EE202 pKa = 4.32EE203 pKa = 4.15IEE205 pKa = 4.01EE206 pKa = 4.06ASRR209 pKa = 11.84QIVEE213 pKa = 4.9SNPDD217 pKa = 3.17MKK219 pKa = 11.03GFALQAYY226 pKa = 7.7GWLYY230 pKa = 10.57EE231 pKa = 4.13QLLANQGALLMNNDD245 pKa = 3.55NGRR248 pKa = 11.84NGTPTEE254 pKa = 4.09VGFTEE259 pKa = 4.96AEE261 pKa = 4.35GKK263 pKa = 10.06SVFAWVEE270 pKa = 3.66RR271 pKa = 11.84MIEE274 pKa = 4.13DD275 pKa = 3.88DD276 pKa = 3.41TFANYY281 pKa = 7.44GTNSDD286 pKa = 3.57NMVSGFMSGDD296 pKa = 2.87VAMFLQSSASVKK308 pKa = 10.49NVVEE312 pKa = 3.97NAPFEE317 pKa = 4.42VGVAFLPYY325 pKa = 10.08PEE327 pKa = 3.93NTEE330 pKa = 3.96RR331 pKa = 11.84NGVVIGGASLWMVDD345 pKa = 3.69GKK347 pKa = 10.26PEE349 pKa = 4.18AEE351 pKa = 5.5QMAAWDD357 pKa = 4.16FMKK360 pKa = 10.5FLQTPEE366 pKa = 4.39VQAQWHH372 pKa = 5.97VGTGYY377 pKa = 10.49FAINPAAYY385 pKa = 9.91DD386 pKa = 3.67EE387 pKa = 4.46EE388 pKa = 4.9VVLEE392 pKa = 4.67AYY394 pKa = 7.67EE395 pKa = 4.14TMPQLKK401 pKa = 8.25VTVDD405 pKa = 3.2QLQATTSNYY414 pKa = 7.76ATQGAIMDD422 pKa = 5.0MIPEE426 pKa = 4.09ARR428 pKa = 11.84RR429 pKa = 11.84ITEE432 pKa = 3.98TALEE436 pKa = 4.18TVYY439 pKa = 11.26NGGAVDD445 pKa = 3.98DD446 pKa = 4.9TYY448 pKa = 11.73QSVVDD453 pKa = 4.52QINAAIEE460 pKa = 3.89QANRR464 pKa = 11.84ARR466 pKa = 11.84GEE468 pKa = 3.89
Molecular weight: 51.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4QL20|W4QL20_9BACI Uncharacterized protein OS=Bacillus hemicellulosilyticus JCM 9152 OX=1236971 GN=JCM9152_3868 PE=4 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1252832 |
37 |
3284 |
288.3 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.555 ± 0.036 | 0.703 ± 0.011 |
5.281 ± 0.038 | 7.891 ± 0.048 |
4.568 ± 0.036 | 6.54 ± 0.038 |
2.361 ± 0.018 | 7.713 ± 0.036 |
5.94 ± 0.037 | 9.771 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.851 ± 0.023 | 4.272 ± 0.027 |
3.471 ± 0.023 | 4.149 ± 0.029 |
4.336 ± 0.025 | 6.161 ± 0.032 |
5.432 ± 0.027 | 7.166 ± 0.036 |
1.147 ± 0.015 | 3.69 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
