
Pseudobutyrivibrio sp. OR37
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Pseudobutyrivibrio; unclassified Pseudobutyrivibrio
Average proteome isoelectric point is 5.68
Get precalculated fractions of proteins
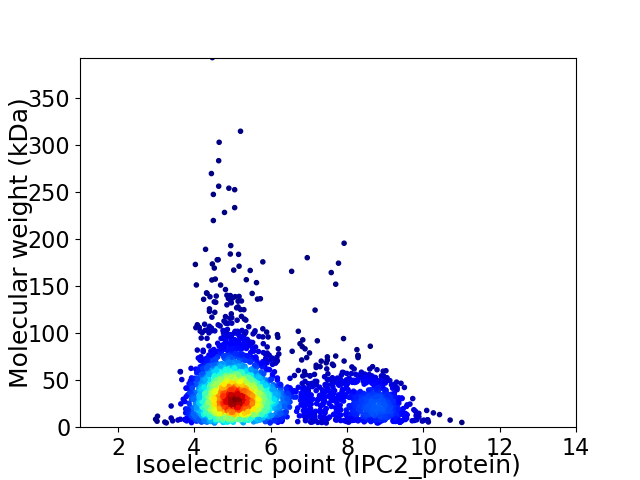
Virtual 2D-PAGE plot for 3127 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3GLN9|A0A1I3GLN9_9FIRM Type I restriction enzyme S subunit OS=Pseudobutyrivibrio sp. OR37 OX=1798186 GN=SAMN04487830_13326 PE=4 SV=1
MM1 pKa = 6.99QNKK4 pKa = 9.24IVTKK8 pKa = 10.65LLTCALSAGLILSGCGNTSVSSTEE32 pKa = 4.11TEE34 pKa = 4.26VTDD37 pKa = 4.48SANEE41 pKa = 3.92AAGSTTIGQTVSGPARR57 pKa = 11.84DD58 pKa = 4.54DD59 pKa = 3.23ISVNEE64 pKa = 4.3TIEE67 pKa = 4.0NNEE70 pKa = 4.34DD71 pKa = 3.27GEE73 pKa = 4.46HH74 pKa = 6.62AIEE77 pKa = 5.23ASGEE81 pKa = 3.96DD82 pKa = 3.13ASYY85 pKa = 11.84SNVEE89 pKa = 4.2VIKK92 pKa = 10.09TGDD95 pKa = 3.33SDD97 pKa = 3.99EE98 pKa = 4.79GDD100 pKa = 3.38EE101 pKa = 4.97ADD103 pKa = 4.87FYY105 pKa = 11.61GDD107 pKa = 3.1NAAIFATDD115 pKa = 3.59GATLDD120 pKa = 3.62LSEE123 pKa = 5.15IVVDD127 pKa = 4.32TNGTHH132 pKa = 6.62ANDD135 pKa = 3.11VFSYY139 pKa = 11.63GEE141 pKa = 4.15GTTVNISDD149 pKa = 4.63SYY151 pKa = 10.16ITTAGNCSGGLMTTGGGTMNATNLTIEE178 pKa = 4.19TSGNSSAAIRR188 pKa = 11.84SDD190 pKa = 3.19RR191 pKa = 11.84GGGTVTVTGGDD202 pKa = 3.76YY203 pKa = 7.6TTSGTGSPVIYY214 pKa = 9.33STADD218 pKa = 3.25VTVSNATLTSTASQGVVVEE237 pKa = 4.67GNNSATLNNVVLNADD252 pKa = 3.87NNTKK256 pKa = 10.61NSDD259 pKa = 3.12KK260 pKa = 10.85SDD262 pKa = 3.51YY263 pKa = 8.76YY264 pKa = 9.41QAVMIYY270 pKa = 10.65QSMSGDD276 pKa = 3.54ADD278 pKa = 3.46EE279 pKa = 5.1GTASFSLKK287 pKa = 10.65DD288 pKa = 3.55SLLNNANGDD297 pKa = 3.74VFFITNTVADD307 pKa = 3.87INIEE311 pKa = 3.82NSEE314 pKa = 4.18INNNGDD320 pKa = 3.84GILLRR325 pKa = 11.84AAAAGWGNEE334 pKa = 3.95GANGGQVNVNVKK346 pKa = 9.53NQSLEE351 pKa = 3.77GDD353 pKa = 3.55MSIDD357 pKa = 4.08DD358 pKa = 3.86VSAMNLYY365 pKa = 10.32LGEE368 pKa = 4.2GATYY372 pKa = 10.62SGAINTNGEE381 pKa = 3.93AGDD384 pKa = 4.03VYY386 pKa = 10.98IQLTDD391 pKa = 3.37GATWTLTGDD400 pKa = 3.78SYY402 pKa = 11.12VTALACDD409 pKa = 3.73ADD411 pKa = 4.45SINLNGYY418 pKa = 8.34KK419 pKa = 10.22LYY421 pKa = 11.41VNGEE425 pKa = 4.28EE426 pKa = 4.09YY427 pKa = 10.73TEE429 pKa = 4.67GSEE432 pKa = 4.31STGDD436 pKa = 3.9AITLEE441 pKa = 4.54KK442 pKa = 9.8ATSSGAPDD450 pKa = 3.2GGMPEE455 pKa = 4.24GKK457 pKa = 9.97APSGEE462 pKa = 4.15KK463 pKa = 10.38PEE465 pKa = 4.46GNPPSGEE472 pKa = 4.12KK473 pKa = 9.88PAGEE477 pKa = 4.27APSGEE482 pKa = 4.42KK483 pKa = 10.42PEE485 pKa = 4.2GTPPSNEE492 pKa = 3.73NSNKK496 pKa = 9.95NN497 pKa = 3.23
MM1 pKa = 6.99QNKK4 pKa = 9.24IVTKK8 pKa = 10.65LLTCALSAGLILSGCGNTSVSSTEE32 pKa = 4.11TEE34 pKa = 4.26VTDD37 pKa = 4.48SANEE41 pKa = 3.92AAGSTTIGQTVSGPARR57 pKa = 11.84DD58 pKa = 4.54DD59 pKa = 3.23ISVNEE64 pKa = 4.3TIEE67 pKa = 4.0NNEE70 pKa = 4.34DD71 pKa = 3.27GEE73 pKa = 4.46HH74 pKa = 6.62AIEE77 pKa = 5.23ASGEE81 pKa = 3.96DD82 pKa = 3.13ASYY85 pKa = 11.84SNVEE89 pKa = 4.2VIKK92 pKa = 10.09TGDD95 pKa = 3.33SDD97 pKa = 3.99EE98 pKa = 4.79GDD100 pKa = 3.38EE101 pKa = 4.97ADD103 pKa = 4.87FYY105 pKa = 11.61GDD107 pKa = 3.1NAAIFATDD115 pKa = 3.59GATLDD120 pKa = 3.62LSEE123 pKa = 5.15IVVDD127 pKa = 4.32TNGTHH132 pKa = 6.62ANDD135 pKa = 3.11VFSYY139 pKa = 11.63GEE141 pKa = 4.15GTTVNISDD149 pKa = 4.63SYY151 pKa = 10.16ITTAGNCSGGLMTTGGGTMNATNLTIEE178 pKa = 4.19TSGNSSAAIRR188 pKa = 11.84SDD190 pKa = 3.19RR191 pKa = 11.84GGGTVTVTGGDD202 pKa = 3.76YY203 pKa = 7.6TTSGTGSPVIYY214 pKa = 9.33STADD218 pKa = 3.25VTVSNATLTSTASQGVVVEE237 pKa = 4.67GNNSATLNNVVLNADD252 pKa = 3.87NNTKK256 pKa = 10.61NSDD259 pKa = 3.12KK260 pKa = 10.85SDD262 pKa = 3.51YY263 pKa = 8.76YY264 pKa = 9.41QAVMIYY270 pKa = 10.65QSMSGDD276 pKa = 3.54ADD278 pKa = 3.46EE279 pKa = 5.1GTASFSLKK287 pKa = 10.65DD288 pKa = 3.55SLLNNANGDD297 pKa = 3.74VFFITNTVADD307 pKa = 3.87INIEE311 pKa = 3.82NSEE314 pKa = 4.18INNNGDD320 pKa = 3.84GILLRR325 pKa = 11.84AAAAGWGNEE334 pKa = 3.95GANGGQVNVNVKK346 pKa = 9.53NQSLEE351 pKa = 3.77GDD353 pKa = 3.55MSIDD357 pKa = 4.08DD358 pKa = 3.86VSAMNLYY365 pKa = 10.32LGEE368 pKa = 4.2GATYY372 pKa = 10.62SGAINTNGEE381 pKa = 3.93AGDD384 pKa = 4.03VYY386 pKa = 10.98IQLTDD391 pKa = 3.37GATWTLTGDD400 pKa = 3.78SYY402 pKa = 11.12VTALACDD409 pKa = 3.73ADD411 pKa = 4.45SINLNGYY418 pKa = 8.34KK419 pKa = 10.22LYY421 pKa = 11.41VNGEE425 pKa = 4.28EE426 pKa = 4.09YY427 pKa = 10.73TEE429 pKa = 4.67GSEE432 pKa = 4.31STGDD436 pKa = 3.9AITLEE441 pKa = 4.54KK442 pKa = 9.8ATSSGAPDD450 pKa = 3.2GGMPEE455 pKa = 4.24GKK457 pKa = 9.97APSGEE462 pKa = 4.15KK463 pKa = 10.38PEE465 pKa = 4.46GNPPSGEE472 pKa = 4.12KK473 pKa = 9.88PAGEE477 pKa = 4.27APSGEE482 pKa = 4.42KK483 pKa = 10.42PEE485 pKa = 4.2GTPPSNEE492 pKa = 3.73NSNKK496 pKa = 9.95NN497 pKa = 3.23
Molecular weight: 50.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3D4E8|A0A1I3D4E8_9FIRM Uncharacterized protein OS=Pseudobutyrivibrio sp. OR37 OX=1798186 GN=SAMN04487830_10943 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.0VHH16 pKa = 5.93GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.0VHH16 pKa = 5.93GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072131 |
40 |
3574 |
342.9 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.493 ± 0.041 | 1.333 ± 0.019 |
6.51 ± 0.036 | 7.364 ± 0.049 |
4.227 ± 0.033 | 6.791 ± 0.039 |
1.592 ± 0.019 | 7.749 ± 0.046 |
7.212 ± 0.041 | 8.302 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.896 ± 0.027 | 5.172 ± 0.036 |
3.039 ± 0.024 | 2.72 ± 0.024 |
3.547 ± 0.032 | 6.072 ± 0.044 |
5.648 ± 0.058 | 6.958 ± 0.031 |
0.872 ± 0.015 | 4.504 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
